Figure 3: RET structural variants on DNA sequencing and rearrangements on FISH.
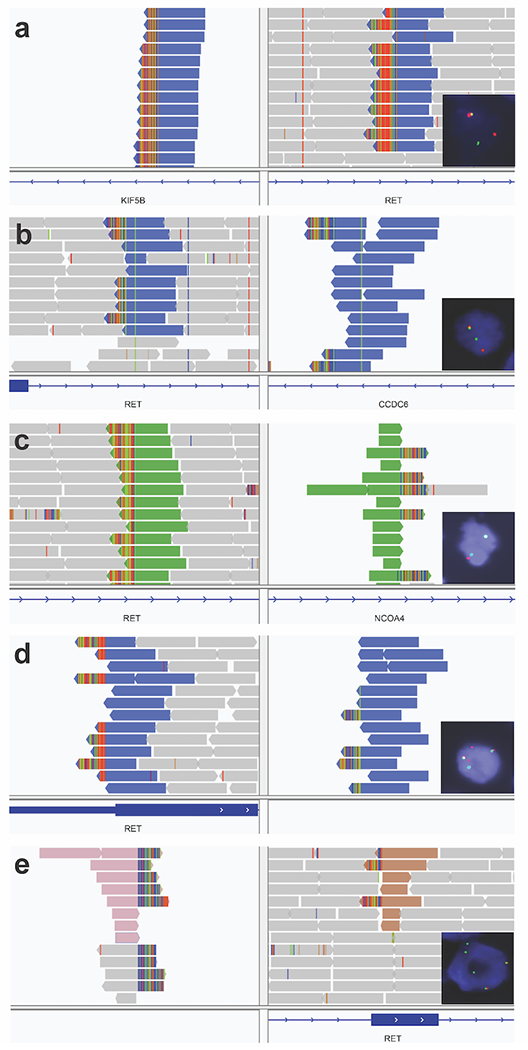
(A) Lung adenocarcinoma (group B) with a KIF5B-RET structural variant (SV). FISH showed a RET rearrangement in 44% of tumor cells. RNA sequencing showed a KIF5B-RET fusion. (B) Anaplastic thyroid carcinoma (group A) with CCDC6-RET SV. FISH showed a RET rearrangement in 68% of tumor cells. RNA sequencing was not performed. (C) Papillary thyroid carcinoma (group A) with NCOA4-RET SV. FISH showed a subtle split pattern that was not considered rearranged. RNA sequencing was not performed. (D) Pancreatic adenocarcinoma (group D) with an inversion connecting an intergenic region (91 Kb before LINC01468) to intron 3 in RET. FISH showed a RET rearrangement in 95% of tumor cells. RNA sequencing did not show a RET fusion. (E) Invasive breast cancer (group D) with a translocation connecting an intergenic region (9178 Kb before PREP) to exon 9 in RET. FISH showed a RET rearrangement in 53% of tumor cells with loss of 5’ signal (red). RNA sequencing did not show a RET fusion.
