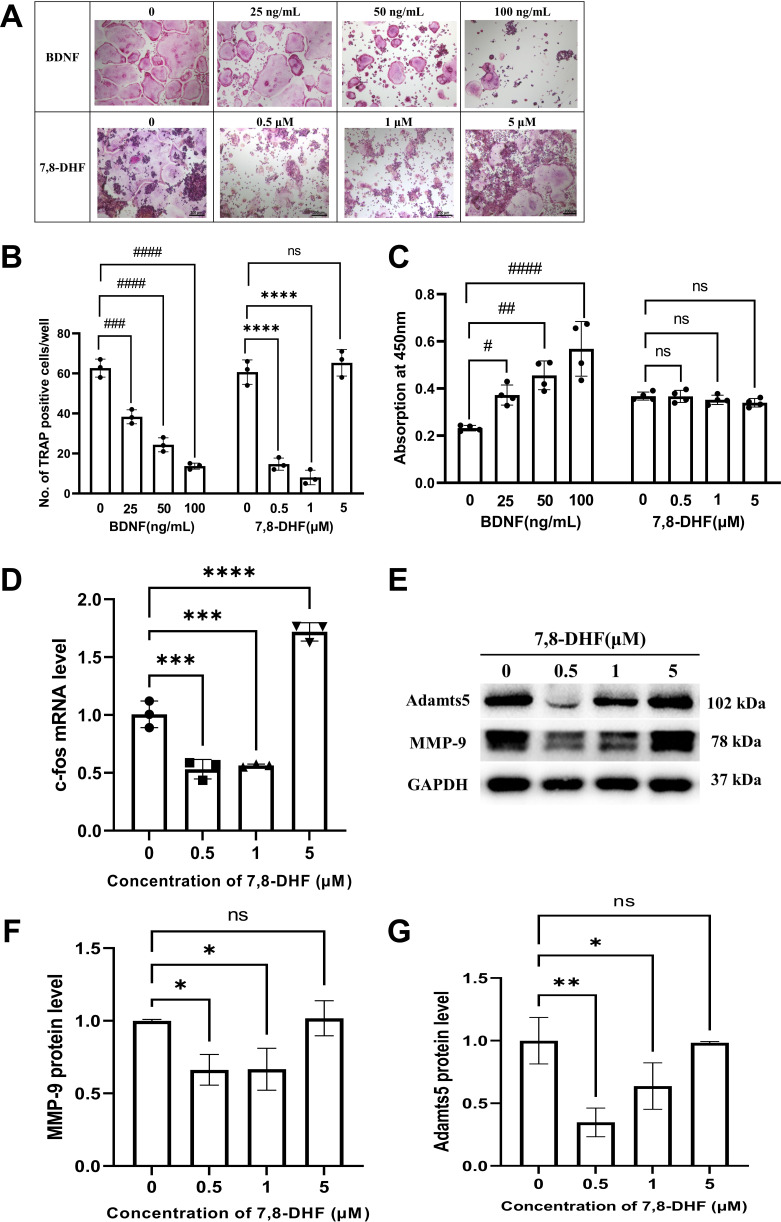
Figure 4. 7,8-Dihydroxyflavone (7,8-DHF) inhibited receptor activator of nuclear factor-κB ligand (RANKL)-induced osteoclastogenesis.
(A) Representative images of tartrate-resistant acid phosphatase (TRAP)-positive multinucleated osteoclasts after the treatment with brain-derived neurotrophic factor (BDNF) or 7,8-DHF for 5 days (magnification: 100×, scale bar: 200 μm). Source files of micrographs used for the quantitative analysis are available in Figure 4—source data 1. (B) The average number of TRAP-positive multinucleated (nuclei ≥ 3) cells per cell. (C) The effects of BDNF or 7,8-DHF on the cytoactive of RAW264.7 cells. (D) The mRNA level of c-fos. Results were normalized to the reference gene GAPDH. (E–G) The protein levels of matrix metalloprotein-9 (MMP-9) and Adamts5. GAPDH was used as an internal control. The expression levels of target proteins in the 0 μM group were normalized to 1. Representative images from three independent experiments are shown in (E). Source files of the full raw unedited blots and blots with the relevant bands labeled were provided in Figure 4—source data 2. All results were expressed as mean ± SD (B, D, F, G: n = 3, C: n=4; #p < 0.05, ##p < 0.01, ###p < 0.001, ####p < 0.0001, ns: not significant, BDNF-treated groups, one-way analysis of variance [ANOVA]; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns: not significant, 7,8-DHF-treated groups, one-way ANOVA).