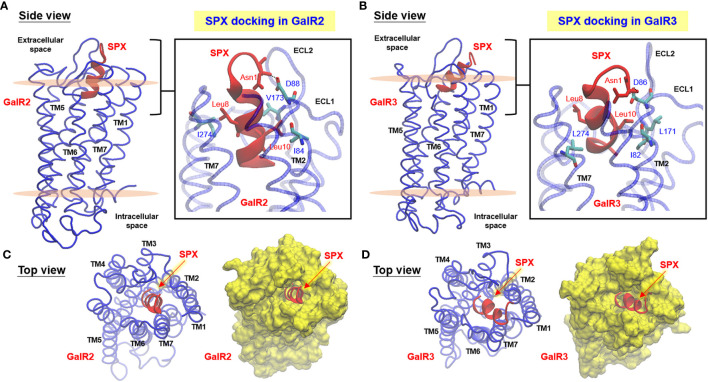
Figure 2.
Docking models for SPX binding with mouse GalR2 and GalR3. Side view of homologous modeling of mouse GalR2 (A) and GalR2 (B) within an asymmetric POPC/POPE lipid bilayer with MD simulation of ligand binding by mouse SPX. Molecular interactions in the well-conserved contact sites in SPX (Asn1, Leu8, and Leu10) with the residues in GalR2 (Ile84, Asp88, Val173, and Ile274) and GalR3 (Ile82, Asp86, Leu171, and Leu274) are presented in the blow-up view close to the surface in the inset of (A, B), respectively. Insertion of the helical region of SPX into the central binding pocket formed by clustering of TMD1–7 of GalR2 (C) and GalR3 (D) could also be noted in the top view of the respective docking models. The helical model of SPX deduced by NMR is marked in red and the corresponding structures for GalR2/3 (including TMD1–7, ECL1–3, and ICL1–3) are presented in blue for the ribbon plot or in yellow for the surface plot of the respective models. Meanwhile, the upper and lower surface of the lipid bilayer in the side view of the docking models are presented in pink color.