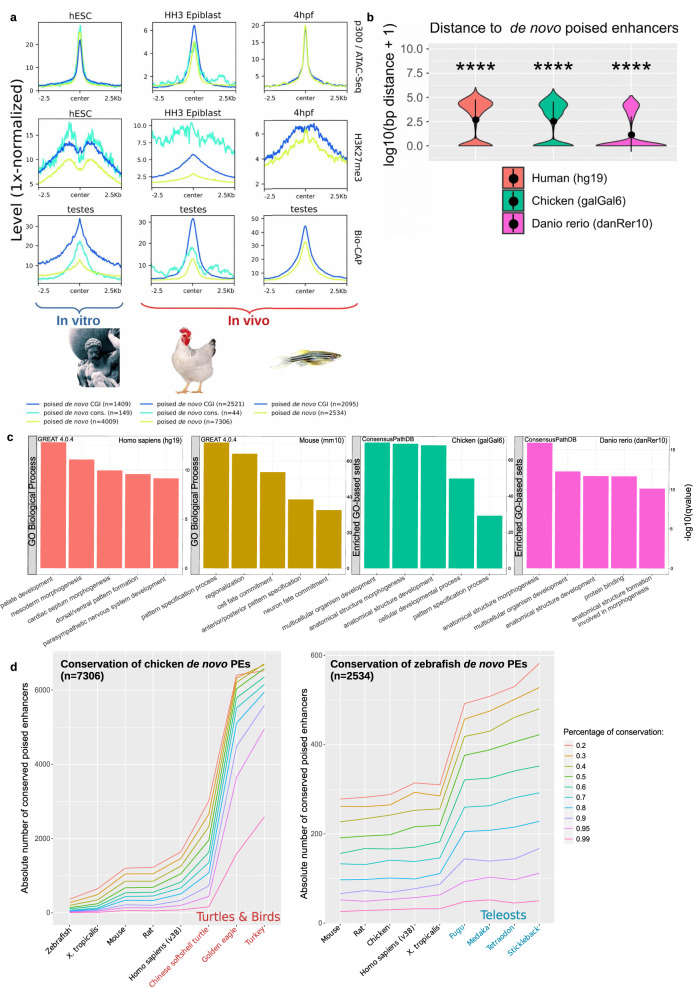
Fig. 2. PEs are a widespread feature across vertebrates.
a PEs were called de novo using available ATAC-Seq, p300 ChIP-seq and H3K27me3 ChIP-Seq data generated in human embryonic stem cells (hESC), HH3 chicken epiblast and zebrafish 4hpf zebrafish embryos (sphere stage; blastulation). ATAC-seq, ChIP-seq, and Bio-CAP6,22 signals are shown for each species around all de novo PEs as well as those overlapping CGIs or previously called conserved mouse PEs (except for zebrafish due to low numbers; n = 2). b Distance between de novo PEs and CGIs for each of the investigated vertebrate species. The asterisks indicate that de novo PEs are significantly closer to CGIs than active mouse enhancers conserved in each species. The exact overlap of de novo PEs (compared to active enhancers) with CGIs increases in all species: in human from 9.85% to 33.18% (p = 6.64e−69; two-sided Wilcoxon test); in chicken from 9.93% to 34.15% (p = 9.88e−27; two-sided Wilcoxon test) and in zebrafish from 2.99% to 68.77% (p = 5.57e−61; two-sided Wilcoxon test). c De novo PE sets were annotated using GREAT 4.0.497 for human and mouse. As this software is not available for chicken and recent zebrafish versions, these de novo PEs were linked to the nearest gene and the resulting gene set was analyzed using ConsensusPathDB98. d The sequence conservation of de novo PEs identified in chicken (left panel) and zebrafish (right panel) was measured across representative species of the main vertebrate clades using different mappability thresholds (mapping ratios 0.2–0.99; color legend). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.