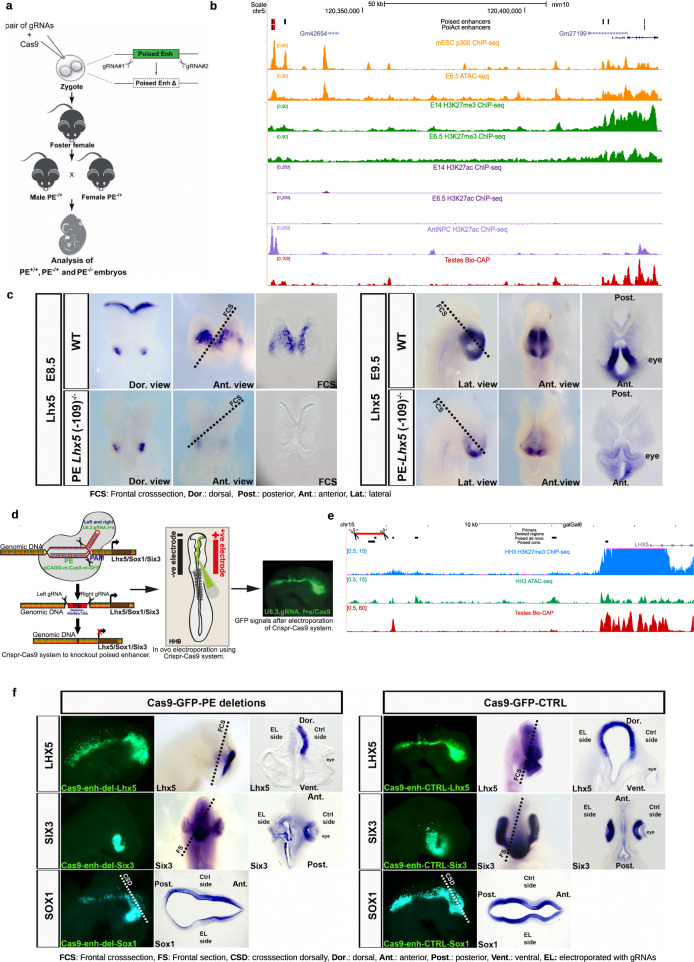
Fig. 6. Functional relevance of PEs during in vivo brain development.
a Graphical overview of the CRISPR/Cas9 experimental strategy used to generate mouse embryos with homozygous deletions of the PE Lhx5 (−109kb). b ATAC-seq and ChIP-seq (p300, H3K27me3, and H3K27ac) profiles generated in mESC and E6.5 mouse epiblast are shown around the Lhx5 locus. In addition, the Bio-CAP profiles generated in mouse testes are shown to illustrate the location of CGIs in the mouse genome, as well as H3K27ac ChIP-seq profiles generated in AntNPC to illustrate the activation of PE Lhx5 (−109 kb) in AntNPC. The genomic region deleted in mouse embryos that includes the PE Lhx5 (−109 kb) is highlighted in red. c RNA in situ hybridizations were performed to visualize Lhx5 expression in WT and PE Lhx5 (−109 kb)−/− mouse embryos at embryonic stages E8.5 (top rows) and E9.5 (bottom rows). Five PE Lhx5 (−109 kb)−/− and four WT mouse embryos were analyzed for each developmental stage, with all PE Lhx5 (−109 kb)−/− embryos showing the reduced Lhx5 expression in the brain as compared to WT. d Graphical overview of the CRISPR/Cas9-based approach used to delete three genetically conserved PEs located within the Lhx5, Sox1, and Six3 loci (left panel). Schematic diagram of in ovo electroporation technique in chick embryo at HH9. Mosaic knockout chicken embryos were generated by unilateral co-electroporation of CAGG > nls-Cas9-nls-GFP and U6.3>PE Lhx5/Sox1/Six3gRNAf + e vectors into the brain region (medium panel). GFP expression in brain and neural tube of an embryo electroporated with the Cas9 and gRNA-GFP vectors (right panel). e ATAC-seq and H3K27me3 ChIP-seq profiles generated in HH3 chicken epiblast are shown around the Lhx5 locus. In addition, Bio-CAP profiles generated in chicken testes are also shown to illustrate the location of CGIs in the chicken genome. The genomic region deleted in chicken embryos that includes the mouse PE Lhx5 (−109 kb) conserved in chicken is highlighted in red. f RNA in situ hybridizations were performed to visualize Lhx5, Six3 or Sox1 expression in HH14-HH16 chicken embryos that were unilaterally electroporated with (i) Left panels: Cas9-GFP and gRNAs flanking the conserved PEs associated with Lhx5, Six3 and Sox1 (Cas9-GFP-PE deletions); (ii) Right panels: Cas9-GFP and scrambled gRNA (Cas9-GFP-CTRL). For Six3, 16 Cas9-GFP-PE deletion embryos were analyzed and all of them showed decreased Six3 expression and an aberrant eye phenotype on the electroporated side. For Lhx5, 20 Cas9-GFP-PE deletion embryos were analyzed and 14/20 embryos showed decreased Lhx5 expression on the electroporated side, while 6/20 embryos showed mild or no reduction. For Sox1, 13 Cas9-GFP-PE deletion embryos were analyzed and 10/13 chicken embryos showed decreased Sox1 expression on the electroporated side, while 3/13 embryos showed no reduction. For each of the previous three genes, we used six Cas9-GFP-CTRL chicken embryos as controls and no changes in gene expression were observed on the electroporated sides.