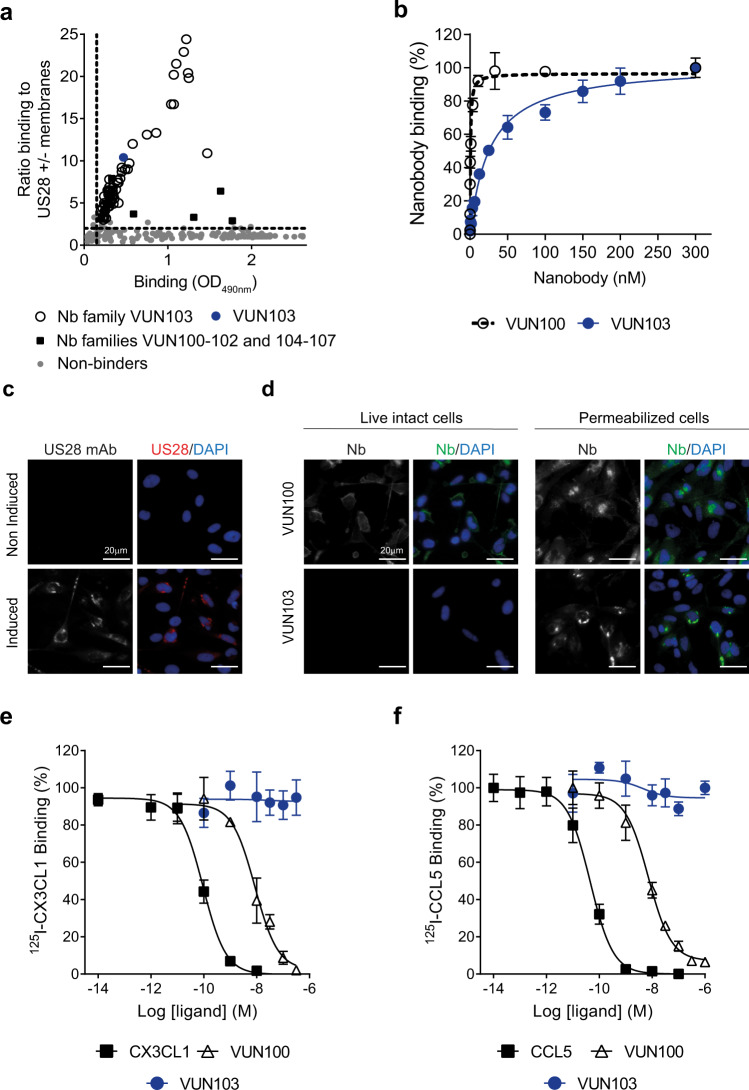
Fig. 1. VUN103 recognizes an intracellular epitope of US28.
a Binding of nanobody-expressing phage, after selection on US28, to US28 measured by phage ELISA. The ratio of binding of phage to membranes with or without US28 was determined and plotted against the total binding of phages to US28-expressing membranes. Data represent one independent experiment. b Binding of the nanobodies VUN100 and VUN103 to US28-expressing membranes, as determined by ELISA. Binding was normalized to binding to US28-negative membranes to determine specific binding (n = 3 independent experiments). c Staining of US28 after 48 h of doxycycline-induction of US28-expressing U251 cells. Non-induced US28-expressing U251 cells served as negative control. Cells were fixed and permeabilized before US28 was visualized using a conventional anti-US28 antibody (US28 mAb; US28, red) and nuclei were stained with DAPI. Representative data of three independent experiments. d Binding of VUN100 and VUN103 to live, intact (non-permeabilized) and fixed and permeabilized US28-expressing U251 cells. For live cells, nanobodies were incubated with cells on ice before cells were fixed. For permeabilized cells, cells were fixed and permeabilized before incubation with nanobodies. Nanobody binding was detected using the Myc-tag and an anti-Myc antibody (Nb, green) and nuclei were stained with DAPI. Representative data of three independent experiments. e, f Displacement of 125I- CX3CL1 (c) and 125I-CCL5 (d) from different US28-expressing membranes by unlabeled ligand (CX3CL1, CCL5, VUN100 or VUN103) (n = 3 independent experiments). All are plotted as mean ± SD. Source data are provided as a Source Data file.