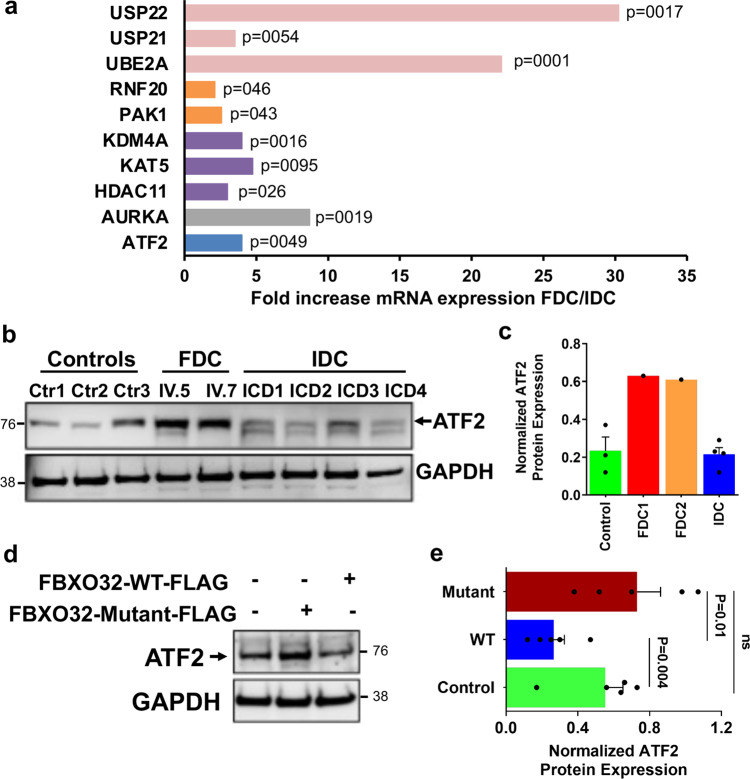
Fig. 4. Increased ATF2 expression in FDC hearts.
a Total RNA was isolated and reverse transcribed from control hearts (n = 3, biological replicates), from the hearts of the patients with the FBXO32 mutation (Family A, n = 4, technical replicates), and from idiopathic dilated hearts (n = 3, biological replicates). mRNA expression for enzymes playing roles in transcription, protein stability, and chromatin regulation was measured by qPCR using the RT2 Profiler PCR Array (Qiagen). Transcripts significantly differentially regulated between the FBXO32 mutant hearts and IDC hearts are shown as fold differences over controls, corrected for 5 housekeeping genes. Significant p values calculated using a Student’s t test are shown. b Western blot analysis performed from human heart lysates prepared from control hearts (n = 3), the two hearts with the FBXO32 mutation (FDC), and idiopathic dilated hearts (IDC) (n = 4) using a specific anti-ATF2 antibody. GAPDH expression was measured in parallel for loading control. c Quantitation of (b) using Adobe photoshop CS6-program. Results are expressed as average ± SD and are normalized against GAPDH. d Representative immunoblots showing ATF2 protein expression in untransfected HEK293 cells or in cells overexpressing mutant-FBXO32 or WT-FBXO32. e Quantitative analysis of (d). Results are normalized against GAPDH and expressed as average ± s.e.m., n = 5 independent experiments. The statistical significance was determined by Student’s t-test. Exact p values are shown. Ns non-significant.