Figure 3.
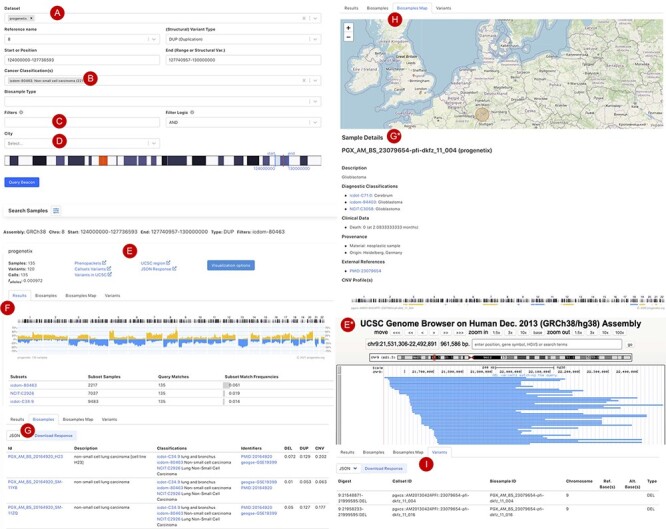
Beacon-style query using fuzzy ranges to identify biosamples with variants matching the CNA range This example queries for a continuous, focal duplication covering the complete MYC gene’s coding region with < = 6 Mb in size. A: Filter for dataset; B: filter for cancer classification (NCIt and ICD-O-3 ontology terms available); C: additional filter, e.g. Cellosaurus; D: additional filter for geographic location; E: external link to UCSC browser to view the alignment of matched variants; F: cancer type classification sorted by frequency of the matched biosamples present in the subset; G: list of matched biosamples with description, statistics and reference. More detailed biosample information can be viewed through ‘id’ link to the sample detail page; H: matched variants with reference to biosamples can be downloaded in json or csv format.
