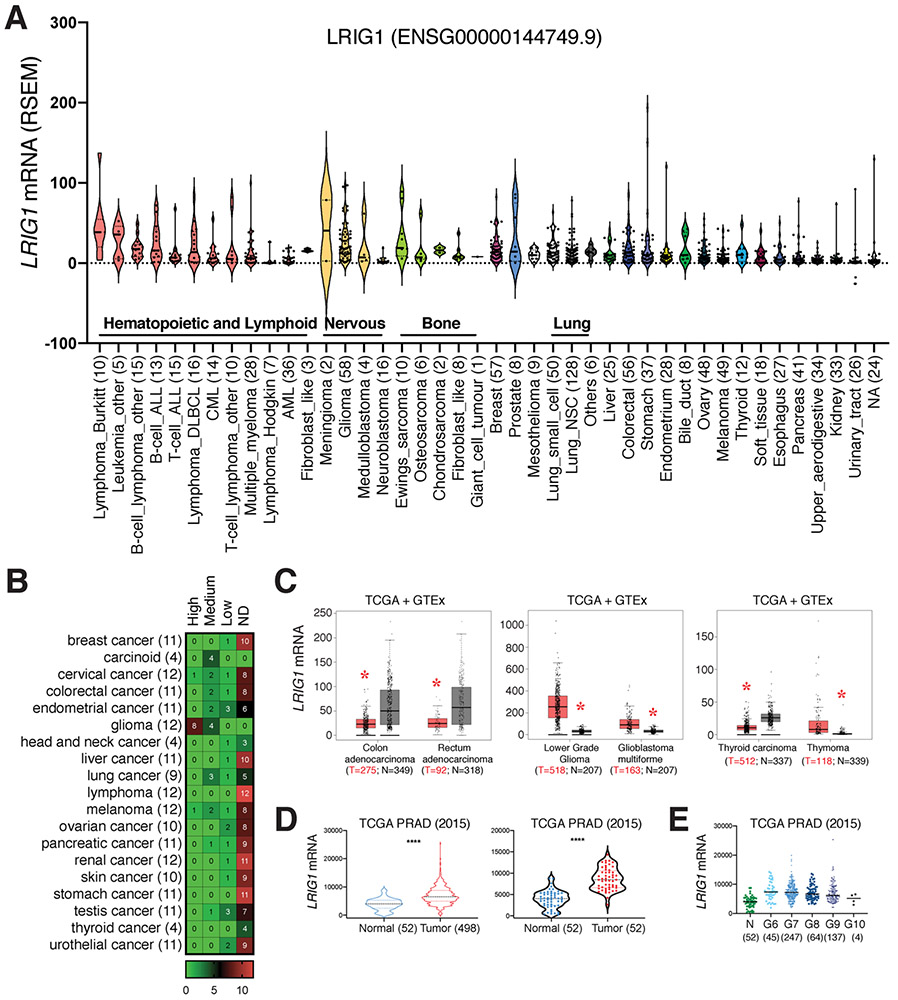
Fig. 2. LRIG1 expression in human cancers and cancer cell lines.
(A) Low levels of LRIG1 mRNA expression in most cultured human cancer cell lines. Gene expression data was extracted from the CCLE Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle). Shown are the violin plots of the LRIG1 mRNA levels (RSEM) in the indicated cancer cell types (n indicated in parentheses). Median and quartiles as indicated. Each dot represents an individual cell line. (B) Heat map of LRIG1 protein expression in the indicated human cancers. The LRIG1 protein expression data was extracted from the Human Protein Atlas, based on immunohistochemistry staining of LRIG1 (HPA011846; see Fig. 1B) in samples from 189 cancer patients of 19 different cancer types (total n indicated in parentheses, scale bar represents case number). ND, not detected. The numbers in individual boxes represent the number of cases. Note that the majority of patient tumors expressed undetectable LRIG1 protein with the glioma as an exception. (C) LRIG1 mRNA expression is decreased in colorectal and thyroid cancers but increased in glioma and thymoma. Shown are the box plots of LRIG1 mRNA levels in tumor samples from TCGA and combined normal tissues from TCGA and GTEx (http://gepia.cancer-pku.cn). *p < 0.05 (ANOVA). (D) Increased LRIG1 mRNA levels in human PCa. Shown are LRIG1 mRNA levels in two different tumor-normal comparisons from the TCGA-PRAD dataset. ****p < 0.0001 (paired Student’s t-test). (E) In TCGA-PRAD dataset, LRIG1 mRNA levels are increased in PCa of all Gleason (G) grades compared to normal (N). p < 0.001 (paired Student’s t-test). However, LRIG1 mRNA levels gradually declined accompanying the increased tumor grade. p < 0.0001 (Jonckheere-Terpstra test).