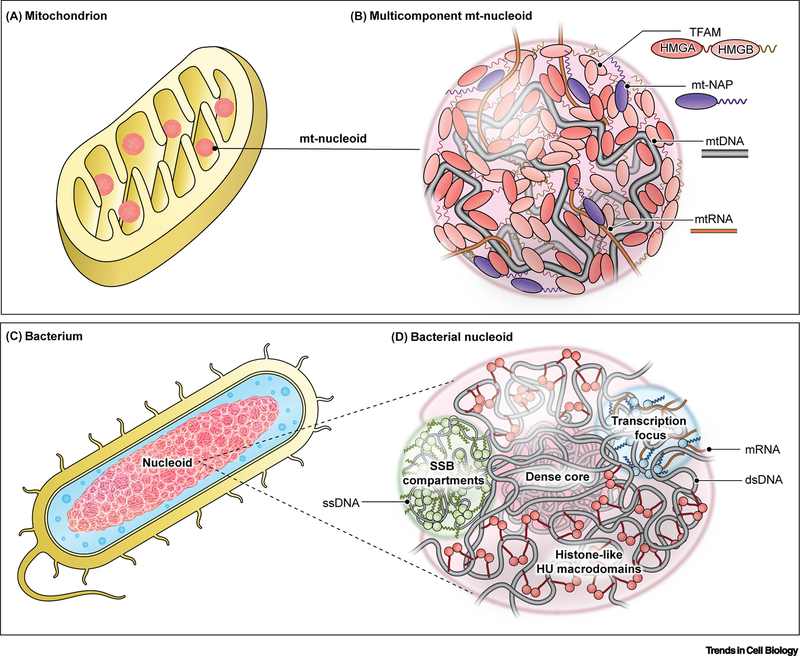
Figure 2. Conservation of phase separation in organization of nucleoids.
(A,B) Phase separation interactions drive the organization of the mitochondrial genome in a mitochondrion (A) into nucleoprotein complexes called mitochondrial nucleoids (B). (B) Mitochondrial Transcription Factor A (TFAM): primary nucleoid architectural protein containing two HMG domains and two IDRs (linker and C-tail); mt-nucleoid associated proteins (mt-NAPs): core proteins associated with mtDNA involved in transcription (TFB2M, POLRMT, mTERF) and replication (mtSSB, POLG1/2, TOP1MT), many of which have DNA-binding domains and intrinsically disordered regions; mtDNA: mitochondrial genome consisting of ∼16 kB circular DNA existing as ∼1–2 copies per nucleoid; mtRNA: mitochondrial RNA transcribed directly from the mitochondrial genome inside the condensate but later associates into separate RNP condensates. (C,D) The larger bacterial genome is analogously organized as a phase-separated nucleoid (D) in a bacterium (C). HU: one of the major architectural, histone-like proteins that exists as a dimer with flexible β-sheet arms that package DNA into a dense core surrounded by less dense phase of DNA and associated proteins; Transcriptional foci: dynamic condensates comprised of RNA polymerase (RNAP) and other transcription factors, many of which contain IDRs; SSB compartments: single-stranded DNA binding protein (SSB) forms a tetramer around single-stranded DNA with protruding intrinsically disordered linkers that are capped by conserved C-terminal peptide motif.