Figure 1. Deregulation of E3-mediated ubiquitylation in cancer.
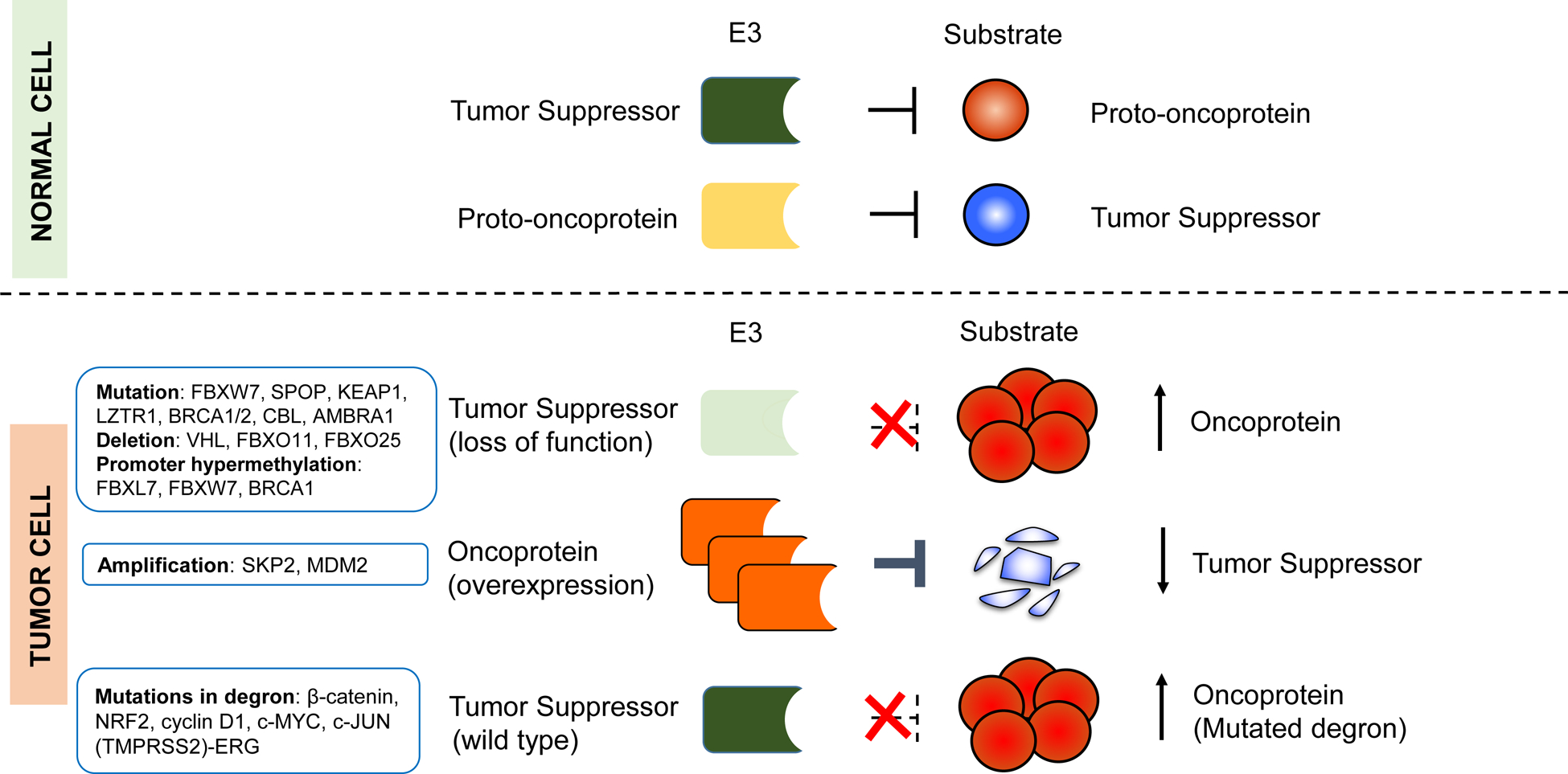
The illustration shows different modes of E3 deregulation, with well-established examples listed. Most deregulations in E3s arise as a consequence of genetic or epigenetic alterations, which can affect the abundance and/or activity of their substrates. For example, deletions, mutations, or promoter methylations can inactivate E3s that normally function as tumor suppressors, and lead to the overexpression of oncoprotein substrates (e.g., c-Myc, cyclin E, and ERG). Alternatively, overexpression of E3s (e.g., through gene amplification of MDM2 and SKP2 loci) that target tumor suppressors (p53 and p27, respectively) can promote tumor formation. In addition, mutations in the substrates, which enable them to escape the recognition by E3s can lead to their accumulation.
