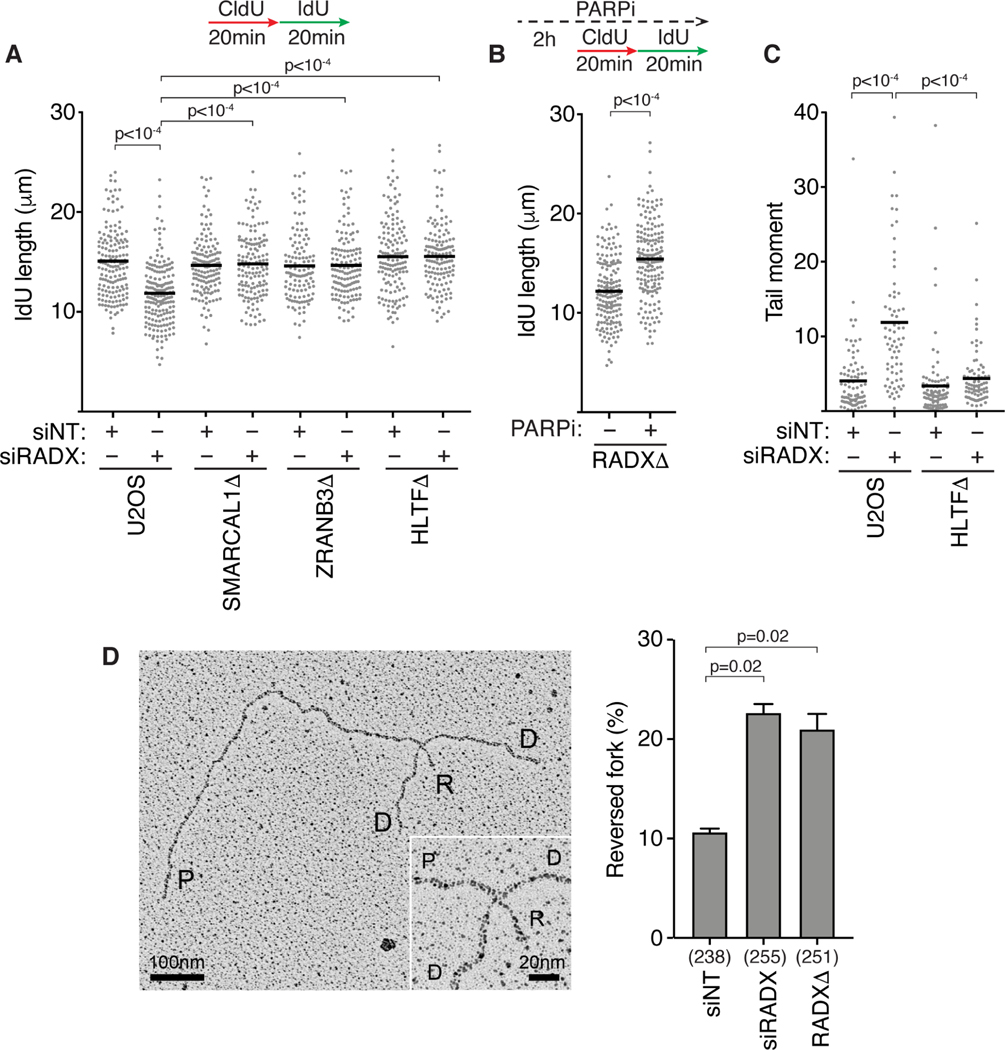
Figure 1. RADX inhibits inappropriate fork reversal in the absence of replication stress.
(A) U2OS cells transfected with the indicated siRNAs (NT=non-targeting) were labeled with CldU followed by IdU and DNA combing was used to measure elongation rates. A one-way ANOVA with Tukey’s multiple comparison test was used to calculate p values in all DNA fiber experiments. (B) Replication fork elongation was monitored by DNA combing in RADXΔ cells treated with 10μM Olaparib as indicated. (C) DSBs were measured by neutral comet assay in wild-type or HLTFΔ U2OS cells transfected with the indicated siRNAs. A Kruskal-Wallis test was used to calculate p values in all comet assays. All fiber and comet assays are representative experiments of at least n=3 biological replicates. See also Figure S1. (D) Example of a reversed replication fork imaged by EM and the mean+/−SEM percentage of reversed forks from three experiments is shown (Inset, magnified four-way junction at the reversed fork; P, parental strands; D, daughter strands; R, reversed strands). The number of replication intermediates analyzed for each condition is indicated in parenthesis. A Welch’s test was used to calculate p values. See also Table S1 and Figure S2.