Abstract
Convolutional neural networks (CNNs) have recently been popular for classification and segmentation through numerous network architectures offering a substantial performance improvement. Their value has been particularly appreciated in the domain of biomedical applications, where even a small improvement in the predicted segmented region (e.g., a malignancy) compared to the ground truth can potentially lead to better diagnosis or treatment planning. Here, we introduce a novel architecture, namely the Overall Convolutional Network (O-Net), which takes advantage of different pooling levels and convolutional layers to extract more deeper local and containing global context. Our quantitative results on 2D images from two distinct datasets show that O-Net can achieve a higher dice coefficient when compared to either a U-Net or a Pyramid Scene Parsing Net. We also look into the stability of results for training and validation sets which can show the robustness of model compared with new datasets. In addition to comparison to the decoder, we use different encoders including simple, VGG Net, and ResNet. The ResNet encoder could help to improve the results in most of the cases.
Keywords: Deep learning, Segmentation, Biomedical imaging
1. Introduction
Training a Convolutional neural networks (CNNs) usually needs thousands of images that can be beyond reach in biomedical applications [1,2]. A sliding-window pattern (patch processing) [3] can make a network to be quite slow, and it is prone to localization accuracy. Therefore, more advanced CNNs are developed to address these issues [4,5]. Two of the frequently used CNNs are U-Net [1] and Pyramid Scene Parsing (PSP) Net [6]. PSP Net uses parallel layers to pool information with different resolutions. This network combines local and global clues to make the final prediction more reliable. However, it can miss information by not extracting higher-order features using deeper layers. On the other hand, U-Net is based on highly deep layers providing depth information from an image. It uses successive layers, where upsampling operators replace pooling operators. These successive layers can increase the resolution of the final outcome while high-resolution features from the contracting path are combined with the upsampled output to increase localization int the final output.
For general segmentation applications, the PASCAL VOC dataset has been considered as one of the best resources for segmentation [7], while the COCO dataset is presented to help instance segmentation [8]. COCO leads to many advanced general segmentation methods [9]. For biomedical images, considering different layers connections to have various segmentation patterns [5], adding residual layers to extract deeper information [10], applying different pooling values to maximize the global context [6], and patch searching in an image [11] are some of the proposed approaches to improve the performance for segmentation. Furthermore, a fully 3D segmentation algorithm has been proposed based on 2D slices and U-Net [12] while V-net [13], which is based on 3D-variant of U-Net analyzed images based on 3D convolutional filters. A combination of DL method and distance regularized level set (DLRS) is proposed to segment lung in computed tomography (CT) images [14]. Breast [15,16] and brain [17,18] cancers have been subjects undergoing intense studies.
In this paper, we present a new CNN architecture for semantic segmentation; specifically designed for biomedical segmentation tasks. Our proposed architecture is built upon the advantages of PSP Net, which has been used frequently for various applications. We add more local context to enrich the information required for final segmentation by considering different encoders and improving the decoder side. The O-Net decoder uses different convolutional and pooling kernels to acquire deeper information from images. The novel contributions of the hereby proposed work are two-fold. First, our effective modification by enriching the local context improves PSP Net and U-Net performances for biomedical tasks. Accordingly, we evaluate the performance of our proposed method compared to two well-established segmentation architectures. Each of these three networks are trained with simple encoder, VGG Net encoder [19], and ResNet encoder [20]. These encoders can add residual or more in-depth information from an image that can help segmentation. Therefore, a total of nine networks are compared here. Our method performs favorably against state-of-the-art methods. Second, we train the CNNs from scratch, do not use already trained model, and do not split train in “pre-train” and “fine-tune” stages. We also consider the complexity of task in our work to evaluate the performance of our method for different tasks. Finally, our code is publicly available through GitHub.
2. Proposed Methods
2.1. O-Net Architecture
A CNN for segmentation contains three major components: encoder, decoder sides, and decision making layers. The encoder layers extract features while the decoder layers try to map the extracted features to pixels level of an input image. The decision-making layers can combine different layers and apply the final decision for the segmentation task based on the extracted features. In addition, training parameters and data augmentation can significantly affect the results. Therefore, in this section, we introduce our proposed network components following by the parameters being used in training. Finally, we briefly discuss the methods used for comparison and the experimental design.
2.2. Encoder Side
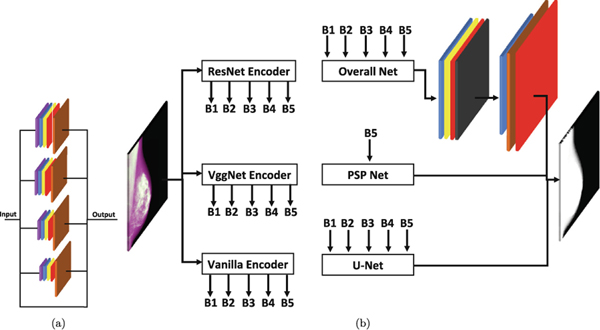
We consider three convolutional or encoder side, encoder, for each network (O-Net, U-Net, and PSP Net). Simple encoder, VGG Net encoder [19], and ResNet encoder [20]. The differences between these three encoders can be seen in Fig. 1. Each of these encoders generates five outputs, referred to as branches (B) as B1, B2, B3, B4, and B5, which is used at different levels of the decoder side.
Fig. 1.
O-Net decoder and other CNNs architectures are shown in this figure. The O-Net decoder layers are illustrated in (a). The brown layer shows the resizing function to return the convoluted result to the original image size. It combines different pooling layers inspired by PSP Net. Detailed information on how the CNNs are connected using encoder and decoder is shown in (b). The top right side of panel (b) provide details about the decision making layer for O-Net.
For the simple encoder, the pooling layers size is (2, 2) while the convolutional layers have a kernel of (3, 3). The filter size for the convolutional layers starts from 64 and increases by a factor of 2 when going to the next layer (deeper to the left). Only, the fifth layer has the same number as the fourth layer (64 × 8 = 1024).
The VGG Net encoder employs the same pooling size of (2, 2) and the same kernel of (3, 3). The filter size has the exact same pattern as described for the simple encoder. Finally, the ResNet encoder has five branches with various kernels. ResNet consists of identity and convolutional blocks. The number of the convolutional block is one for all of the branches (after the first branch). However, second, third, fourth, and fifth branches have 2, 3, 4, and 2 identity blocks. Figure 1 shows more details about two of these branches (the second branch has two identity blocks). More details can be found in the code.
2.3. Decoder Side
The de-convolutional or decoder side of O-Net takes advantage of different pooling layers being inspired by PSP Net. We use different pooling layers and combine the results with the original input to mix the local and global context. The original input for each layer helps to improve the localization of the outcome. This decoder module has been illustrated in Fig. 1.
The decoder module includes five divisions with four different pooling kernels (1, 2, 3, and 6) and one layer which passes the input to improve the final localization. Each layer has one average pooling layer, one convolutional layer, one batch normalization layer, and one activation layer. The convolutional layer is designed by a kernel size of (1, 1) and 512 filters. These layers are followed by a resizing component to return all the images to the original input size.
2.4. Decision Making Layers
As explained in Sects.2.2 and 2.3, five branches from one of the three encoders are the input of the decoder module. The output of the decoder module is five branches. We combine these branches and apply a convolutional step (one convolutional layer, one batch normalization, and one activation layer), as shown in Fig. 1. The convolutional layer has a kernel size of (3, 3) with 512 filters.
We use a drop out layer to reduce the chance of overfitting. Finally, another convolutional layer following by the image resizing component is used to have the original image resolution in output. Finally, an activation layer with the softmax function is used to differentiate the segmentation classes.
2.5. Methods for Comparison
We compare our method with U-Net [1] and PSP Net [6]. How these two networks and O-Net are used in this study have been shown in Fig. 1. O-Net and U-Net use all five branches generated by the encoder side while the PSP Net uses the last branch (B5) from each of the encoders. Therefore, we have a total of nine CNNs (three networks and three encoders) for comparison.
We use Adam optimizer with a learning rate of 1e−4 for all nine CNNs.
3. Experimental Setup
3.1. Datasets
In this work, we use two datasets: breast and brain scans. The breast dataset is in gray-scale (one channel) while for the brain dataset we considered gray-scale (one channel) and color images (three channels) to make different complexity levels. All of the images are resized to 256 × 256 pixels. Therefore, a total of three image sets are used in this study.
Gray-scale images for breast tissue segmentation: Segmentation of breast and calculating breast density percentage can is helpful for assessing breast cancer risk [21]. The breast dataset consists of 1,100 digital mammography images from 1,100 patients. 550 images are screened from left and the other half from right breast at the Hospital of the University of Pennsylvania between 2010 and 2014. Women were imaged per U.S. Food and Drug Administration approved protocol consisting of full-field digital mammography in both mediolateral oblique views using Selenia Dimensions, Hologic Inc. The breast area is manually differentiated from the background and pectoralis muscle using ImageJ software [22]. The background segmentation can be considered as a simple task while the pectoralis muscle can be challenging. This dataset includes gray-scale normalized images with 8-bit from the digital mammography images. We use half of the images to train and the other half to test the model. This dataset has two classes as breast and a combination of background and pectoralis muscle.
Gray-scale images for brain tumor segmentation: The brain dataset we used in this study describes the BraTS [17], [18] component of the publicly available Medical Segmentation Decathlon dataset [23]. The training dataset, which has images and labels, consists of four channels: 1) Fluid-attenuated inversion recovery (FLAIR), 2) native T1 weighted, 3) T1 post-contrast, and 4) T2-weighted MRI scans. The BraTS training set has 3D images from 484 patients with four classes of segmentation comprising 1) background, 2) edema, 3) non-enhancing tumor, and 4) enhancing tumor. Since the brain scans were 3D, we decided to extract 2D slices from each patient and randomly select five of them that encompass at least two out of the four segmentation classes. Therefore, we generate 1,200 images for training and 736 images for testing. In order to have a gray-scale, we considered the first channel (FLAIR).
Color Images for Brain Tumor Segmentation: We use the same BraTS dataset with the same number of images. However, we add two other channels (T1 and T1gd) to make color images.
3.2. Metric in Training and Measures for Comparisons
The segmentation performance is measured using two parameters: 1) dice similarity coefficient, and 2) dice weighted. We extend dice formula considering weights for each class based on the number of pixels for that class.
4. Results
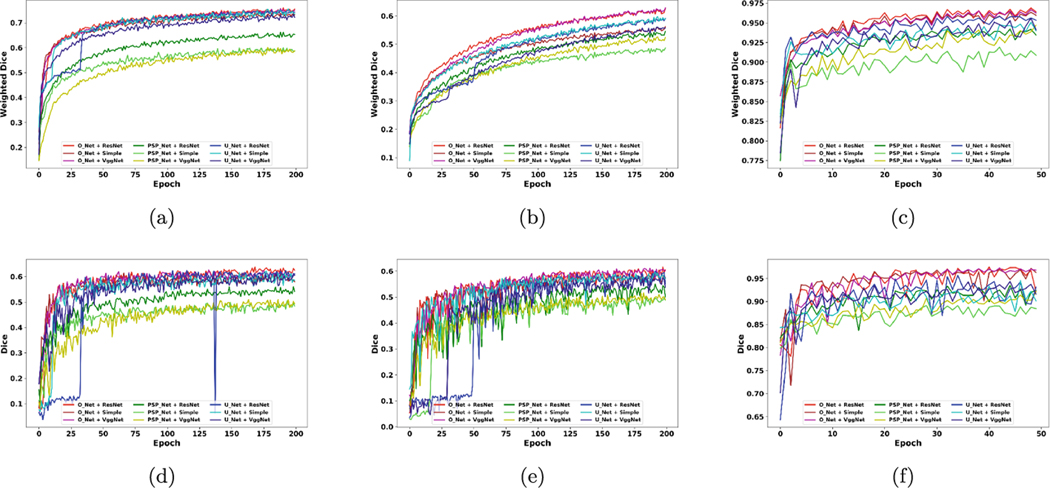
This section presents comparisons between the nine CNNs using the three datasets mentioned in Sect.3.1. All networks are trained using the dice weighted which is formulated in Sect.3.2. We show training and testing sets results for all epochs for all training epochs, as illustrated in Fig. 2. The results for training and testing sets are compared using weighted dice and dice, respectively. The CNNs are trained till they show some signs of overfitting, e.g., Fig. 2 (b) depicts an increase while (d) reaches a plateau, or they reach a plateau for dice weighted, e.g., Fig. 2 (c) and (f). Therefore, using this strategy, we train the brain networks up to 200 epochs and the breast networks up to 50 epochs. The main reason for considering such a condition in training is that we can compare the top and bottom half of training epochs to visualize any possible patterns in the CNNs taring and testing phases.
Fig. 2.
The results for training and testing. Plots in (a), (b), and (c) show the training results for all epochs (200 for BRATS related datasets and 50 for the breast dataset). Panels in (d), (e), and (f) are the testing results corresponding to the training epochs. Each color shows one of the nine CNNs. Red, brown, and purple show the results for O-Net with ResNet, simple, and VGG Net encoders, respectively. Dark green, bright green, and yellow show the results for PSP Net with ResNet, simple, and VGG Net encoders, respectively. Bright blue, cyan, and dark blue show the results for U-Net with ResNet, simple, and VGG Net encoders, respectively. The color brain dataset results are shown in (a) and (d). The gray-scale brain dataset results are shown in (b) and (e). Finally, the breast dataset results are illustrated in (c) and (f).
It should be noted that we use the train and test sets (no validation set used for selecting an epoch). The reason is that we study the performance of networks during the whole training phases on our testing set without selecting one specific epoch. A stable network can be more reliable when an epoch is selected based on a measure on a validation set as the network shows fewer variations and is expected to perform the same on a test set. An unstable network can be highly dependent on the selected epoch and the dataset.
The comparisons are performed by combining the results for the CNNs based on two categories: 1) network-based comparison showing the results for all nine networks; 2) decoder-based and encoder-based comparisons illustrating the results for three decoders (O-Net, PSP Net, and U-Net) and three encoders (simple, VGG Net, and ResNet); In addition, we separate the analysis for the first and second half of the training epochs. This can help us to study a network performance while there is so much information to learn (the first half of training phase, referred to as head) and while a network is almost close to the plateau of learning (the second half of training phase, referred to as tail). In other words, this type of comparison can show which network is more stable in the results. Therefore, the results can be studied as follow:
Networks-based Comparison: Table 1, which summarizes the testing set results in Fig. 2, illustrates the average and standard division of trained nine networks for the head and tail parts of training. O-Net achieved its best performance using VGG Net for the head and tail portion of the training. VGG Net shows about one percent higher dice measure for four out of six conditions (conditions are made by three datasets and two training phases). The ResNet encoder is usually the second network.
Decoder-based and Encoder-based Comparisons: A comprehensive comparison for the effect of decoders is performed and reported in Table 2. The results indicate that O-Net decoder remains on top of all conditions. An almost five percentage difference between O-Net and U-Net for the tail of the breast dataset might suggest the advantages of our network. In addition, Table 2 illustrates the results for encoder side of CNNs. The results show an average of better performance for ResNet (about one percent) while the simple encoder showed better performance at the earlier stage.
Table 1.
The dice results for all nine networks are summarized here. The name of networks shows encoder + decoder. The head and tail depict the results for the first and second half portions of training epochs. The best network is colored in blue for datasets (three datasets) and training phases (head and tail).
| Networks | Head | Tail | ||||
|---|---|---|---|---|---|---|
| Brain Color | Brain Gray | Breast | Brain Color | Brain Gray | Breast | |
| ResNet + O-Net | 54.611.3 | 48.910.5 | 91.45.0 | 61.70.9 | 58.51.7 | 96.01.4 |
| Simple + O-Net | 54.67.9 | 48.79.9 | 92.55.5 | 59.11.3 | 56.71.1 | 95.81.5 |
| VGG Net + O-Net | 55.59.8 | 49.69.6 | 91.44.7 | 60.60.9 | 58.91.1 | 96.30.8 |
| ResNet + PSP Net | 45.67.8 | 41.68.7 | 88.72.8 | 53.60.9 | 51.42.4 | 91.71.1 |
| Simple + PSP Net | 41.87.7 | 35.814.5 | 85.52.1 | 47.61.1 | 47.21.7 | 88.00.9 |
| VGG Net + PSP Net | 38.28.5 | 38.98.5 | 86.42.0 | 48.31.5 | 48.31.7 | 89.71.2 |
| ResNet + U-Net | 41.822.3 | 30.520.6 | 88.96.6 | 60.15.5 | 56.52.1 | 92.42.3 |
| Simple + U-Net | 50.614.6 | 48.98.1 | 88.52.1 | 58.71.5 | 56.91.6 | 91.01.5 |
| VGG Net + U-Net | 50.68.9 | 36.018.4 | 89.24.7 | 58.31.2 | 54.61.7 | 92.21.3 |
Table 2.
A summary by grouping the testing set results based on the decoders and encoders. Amongst the encoders, the simple encoder achieved the best performance for the head phase while ResNet had a higher dice for the tail parts of training phase. The best encoder is colored in green for each dataset. O-Net remains the best encoder. The best decoder is colored in blue for each dataset.
| Name | Head | Tail | |||||
|---|---|---|---|---|---|---|---|
| Brain Color | Brain Gray | Breast | Brain Color | Brain Gray | Breast | ||
| Decoder | O-Net | 54.99.7 | 49.010.0 | 91.85.0 | 60.41.5 | 58.11.7 | 96.11.3 |
| PSP Net | 41.98.5 | 38.711.2 | 86.92.7 | 49.83.0 | 49.02.6 | 89.81.9 | |
| U-Net | 47.616.7 | 38.518.3 | 88.94.8 | 59.03.4 | 56.02.1 | 91.81.8 | |
| Encoder | ResNet | 47.316.0 | 40.316.1 | 89.75.2 | 58.54.8 | 55.53.6 | 93.42.5 |
| Simple | 49.011.8 | 44.412.7 | 88.84.6 | 55.15.5 | 53.64.8 | 91.63.5 | |
| VGG Net | 48.111.6 | 41.514.2 | 89.04.5 | 55.75.5 | 54.04.6 | 92.73.0 | |
5. Conclusions
In this work we introduced a new DL architecture for semantic segmentation, namely O-Net (Fig. 1). The presented CNN is compared with two popular architectures, U-Net and PSP Net. The comparisons are performed by changing the encoder and decoder possibilities. Figure 2 shows the results for such a comparison. In addition, we analyzed the nine CNNs more in detail for the early and late phases of training in Table 1 and Table 2.
We train and present the results for training set based on weighted dice while we report the testing results by dice, as the latter is the most common measure used for all segmentation tasks in the literature. However, it should be noted that the weighted dice had similar patterns with dice on the testing dataset. Figure 2 shows that the O-Net could achieve a higher weighted dice (less loss function) in the training set. This can be more obvious in Fig. 2 (b) and (c).
On the other hand, for the testing set, we analyzed more in depth, shown in Fig. 2, Table 1, and Table 2. We have six conditions based on the training phases (head and tail) and datasets (three datasets). Table 1 and Table 2 are derived from Fig. 2; however, they suggest important notes as follow:
O-Net remains the best network in all six conditions, shown in Table 2.
ResNet shows to be the best encoder in overall, shown in Table 2, especially for the tail portion of training. A reason for such a pattern can be the fact that ResNet tends to go deeper, and it needs a larger number of epochs to converge. However, when it converges it is shown to be more accurate. However, O-Net shows a higher performance for the VGG Net encoder for four out of six conditions. Although the ResNet and VGG Net results are just about one percent different from each other, the reason for such a difference can be the rich feature being extracted in the decoder parts of O-Net.
In the earlier stages of the training, the head phase, we can see that the simple encoder performed slightly better. This can be because of the need for fewer iterations for learning some patterns from images when using the simple encoder compared with the other encoders.
In the earlier stages of the training, the head phase, we can see that the O-Net achieve significantly higher average dice compared to U-Net and PSP Net for all three datasets. This pattern is more obvious in Fig. 2, panels (d) and (e). This suggests that O-Net can reach it’s best performance with fewer iteration.
We can conclude that O-Net with VGG Net encoder can be the best network amongst these nine CNNs. Notably, O-Net seems to be the most stable network on the testing set revealing its reliability. A robust and stable network results on validation set during the entire training phase can be really important because a new variations can cause the network performing worst than our expectations.
One of the limitations for O-Net is the required time for training that might be up to twice of U-Net. Also, saving the weights needs twice more space. In our future works, we intend to extend this work for 3D segmentation tasks as the current version of code is limited solely to 2D. It will let us attend and compare our results with the available challenges, e.g., BraTS. In addition, we plan to evaluate the method for the general segmentation tasks and compare the results with the medical datasets. Last but not least, we can comprehensively investigate the stability of network during the training phase on larger datasets.
Note: Sample images showing segmented area using nine networks and the code can be found in supplementary materials (which will be added on GitHub).
Supplementary Material
Acknowledgments.
Research reported in this publication was partly supported by the National Institutes of Health (NIH) under award numbers NINDS:R01NS042645, NCI: R01CA161749, NCI:U24CA189523, NCI:U01CA242871. The content of this publication is solely the responsibility of the authors and does not represent the official views of the NIH. This work was also supported by the Susan G. Komen for the Cure® Breast Cancer Foundation [PDF17479714]. Also, we appreciate NVIDIA support for a donation of GPU to OHM.
Footnotes
Electronic supplementary material The online version of this chapter (https://doi.org/10.1007/978-3-030-59861-721) contains supplementary material, which is available to authorized users.
References
- 1.Ronneberger O, Fischer P, Brox T: U-net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A. (eds.) MICCAI 2015. LNCS, pp. 234–241. Springer, Heidelberg: (2015). 10.1007/978-3-319-24574-428 [DOI] [Google Scholar]
- 2.Mortazi A, Bagci U: Automatically designing CNN architectures for medical image segmentation. In: Shi Y, Suk H-I, Liu M. (eds.) MLMI 2018. LNCS, vol. 11046, pp. 98–106. Springer, Cham: (2018). 10.1007/978-3-030-00919-912 [DOI] [Google Scholar]
- 3.Ciresan D, Giusti A, Gambardella LM, Schmidhuber J: Deep neural networks segment neuronal membranes in electron microscopy images. In: Advances in Neural Information Processing Systems, pp. 2843–2851 (2012) [Google Scholar]
- 4.Iglovikov V, Shvets A: Ternausnet: U-net with VGG11 encoder pre-trained on imagenet for image segmentation. arXiv preprint arXiv:1801.05746 (2018) [Google Scholar]
- 5.Murugesan B, Sarveswaran K, Shankaranarayana SM, Ram K, Sivaprakasam M: Psi-net: Shape and boundary aware joint multi-task deep network for medical image segmentation. arXiv preprint arXiv:1902.04099 (2019) [DOI] [PubMed] [Google Scholar]
- 6.Zhao H, Shi J, Qi X, Wang X, Jia J: Pyramid scene parsing network. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2881–2890 (2017) [Google Scholar]
- 7.Everingham M, Eslami SMA, Van Gool L, Williams CKI, Winn J, Zisserman A: The Pascal visual object classes challenge: a retrospective. Int. J. Comput. Vis 111(1), 98–136 (2015) [Google Scholar]
- 8.Lin T-Y, et al. : Microsoft COCO: common objects in context. In: Fleet D, Pajdla T, Schiele B, Tuytelaars T. (eds.) ECCV 2014. LNCS, vol. 8693, pp. 740–755. Springer, Cham: (2014). 10.1007/978-3-319-10602-148 [DOI] [Google Scholar]
- 9.Li Y, Qi H, Dai J, Ji X, Wei Y: Fully convolutional instance-aware semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2359–2367 (2017) [Google Scholar]
- 10.Zhu Z, Liu C, Yang D, Yuille A, Xu D: V-NAS: Neural architecture search for volumetric medical image segmentation. arXiv preprint arXiv:1906.02817 (2019) [Google Scholar]
- 11.Liu W, Rabinovich A, Berg AC: Parsenet: Looking wider to see better. arXiv preprint arXiv:1506.04579 (2015) [Google Scholar]
- 12.Çiçek O, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham: (2016). 10.1007/978-3-319-46723-849 [DOI] [Google Scholar]
- 13.Milletari F, et al. : Hough-CNN: deep learning for segmentation of deep brain regions in MRI and ultrasound. Comput. Vis. Image Understand 164, 92–102 (2017) [Google Scholar]
- 14.Ngo TA, Carneiro G: Fully automated segmentation using distance regularized level set and deep-structured learning and inference. In: Lu L, Zheng Y, Carneiro G, Yang L. (eds.) Deep Learning and Convolutional Neural Networks for Medical Image Computing. ACVPR, pp. 197–224. Springer, Cham: (2017). 10.1007/978-3-319-42999-112 [DOI] [Google Scholar]
- 15.Kallenberg M, et al. : Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans. Med. Imaging 35(5), 1322–1331 (2016) [DOI] [PubMed] [Google Scholar]
- 16.Maghsoudi OH, Gastounioti A, Pantalone L, Conant E, Kontos D: Automatic breast segmentation in digital mammography using a convolutional neural network. In: 15th International Workshop on Breast Imaging (IWBI2020), vol. 11513, p. 1151322 (2020) [Google Scholar]
- 17.Bakas S, et al. : Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 4, 170117 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakas S, et al. : Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. arXiv preprint arXiv:1811.02629 (2018) [Google Scholar]
- 19.Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014) [Google Scholar]
- 20.Szegedy C, Ioffe S, Vanhoucke V, Alemi AA: Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-First AAAI Conference on Artificial Intelligence (2017) [Google Scholar]
- 21.Kontos D, et al. : Radiomic phenotypes of mammographic parenchymal complexity: toward augmenting breast density in breast cancer risk assessment. Radiology 290(1), 41–49 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rueden CT, et al. : Imagej 2: Imagej for the next generation of scientific image data. BMC Bioinform. 18(1), 529 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simpson AL, et al. : A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv preprint arXiv:1902.09063 (2019) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.