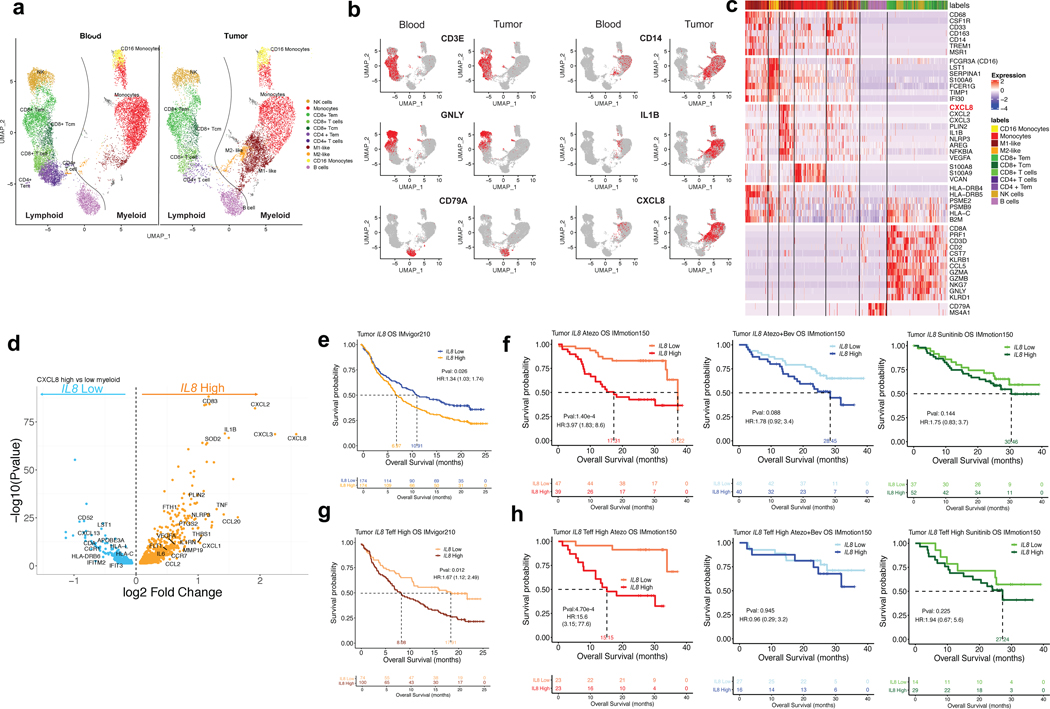
Figure 4. Single-cell RNASeq analysis of IL8 gene expression in immune subsets from matched intratumoral and peripheral blood leukocytes from RCC patients and association of tumor IL8 gene expression with clinical outcomes in mUC and mRCC.
a, Single cell RNAseq (scRNAseq) identifies immune cells subsets in leukocytes in four matched blood and tumor from mRCC. UMAP plot of 25,459 immune cells, color-coded by their annotation from four matched blood and tumor (single cell n= 13694 and 11765 cells, respectively). Color-coded expression (grey to red) of the lymphocytic (left two panels) and myeloid (right two panels) cell type-specific markers from four matched blood and tumor shown in b. c, Heat map reporting scaled expression (log expression count values) of selected gene sets. Gene expression color scheme is based on scale log count expression distribution, from –2.0 (blue) to 2.0 (red). Color bars in right margin highlight cell subsets of interest. d, Differential gene expression analysis between IL8-high and -low intratumoral myeloid cells showing enriched expression of myeloid inflammatory response genes (orange) in IL8 high myeloid cells versus higher expression of antigen presentation genes (blue) in IL8 low myeloid cells. Volcano plot representing differentially expressed (FDR-corrected p<0.05) between IL8 high and IL8 low myeloid cells in the four mRCC tumors (n= 6280 cells). Differential expression analysis with the generalized linear models (glm)-based statistical methods of the edgeR package with Benjamini & Hochberg corrections. e. High vs. low intratumoral IL8 gene expression and overall survival (OS) following atezolizumab monotherapy in IMvigor210 (HR: 1.34, 95% CI:1.03, 1.74, P=0.026). f, Kaplan-Meier curves depict overall survival of IL8 expression in the tumors of IMmotion using median cutoff in atezolizumab, atezolizumab + bevacizumab and sunitinib arms. High vs. low intratumoral IL8 gene expression and OS with atezolizumab monotherapy (HR: 3.97, 95% CI:1.83, 8.6, P=1.40e-4), atezolizumab + bevacizumab (HR: 1.78, 95% CI: 0.92, 3.4, P= 0.088), and sunitinib (HR: 1.75, 95% CI: 0.83, 3.7, P= 0.144) in IMmotion 150. g, High versus low intratumoral IL8 gene expression and OS in Teff-high patients who received atezolizumab monotherapy in IMvigor210 (HR: 1.67, 95% CI: 1.12, 2.49, P=0.012). h, Kaplan-Meier curves depict overall survival of IL8 expression in the tumors of IMmotion using median cutoff in atezolizumab, atezolizumab + bevacizumab and sunitinib arms. High versus low intratumoral IL8 gene expression and OS in the T-effector (Teff) high patient subset treated with: atezolizumab monotherapy (HR: 15.6, 95% CI: 3.15, 77.6, P=4.70e-4), atezolizumab + bevacizumab (HR: 0.96, 95% CI: 0.29, 3.2, P=0.945), and sunitinib (HR: 1.94, 95% CI: 0.67, 5.6, P=0.225) in IMmotion150. HRs in Figure 4e-h were calculated using stratified Cox proportional hazard regression models, and P values were calculated using stratified log-rank test. P values were adjusted for multiple comparisons. Multivariate analyses adjusted HRs for age, sex, race, ECOG performance status, presence of liver metastasis, and tumor burden (sum of longest diameter, SLD) in mUC; and age, sex, Memorial Sloan Kettering Cancer Risk (MSKCC) prognostic risk score, previous nephrectomy, and SLD in mRCC data sets.