Figure 2.
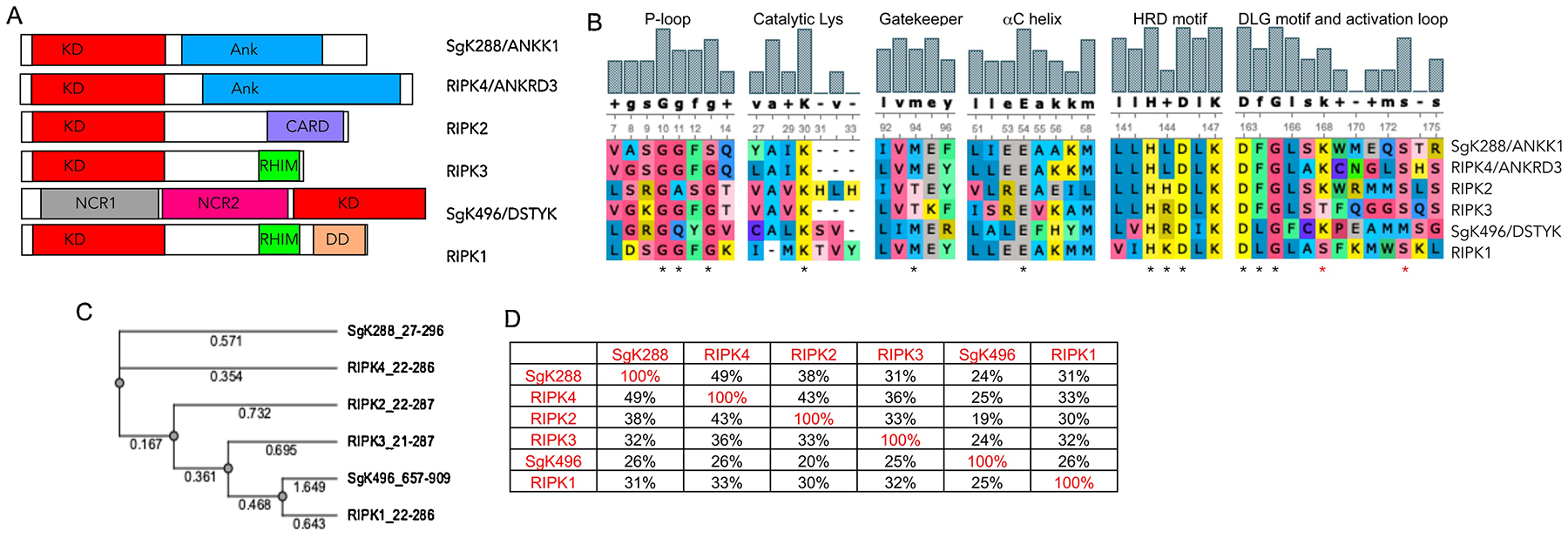
A) Domain architecture of RIPKs. B) Alignment of the critical catalytic elements of RIPKs. * indicate critical residues in each motif, * depict RIPK1 autophosphorylation sites Ser161 and Ser166. C) Phylogenic tree of RIPK kinase domains. Alignments in B and C were generated using UGENE software package and T-Coffee alignment algorithm. D) Degree of similarity between RIPKs was calculated in UGENE using the built-in similarity matrix. Alignment was also performed using ClustalW with similar results.
