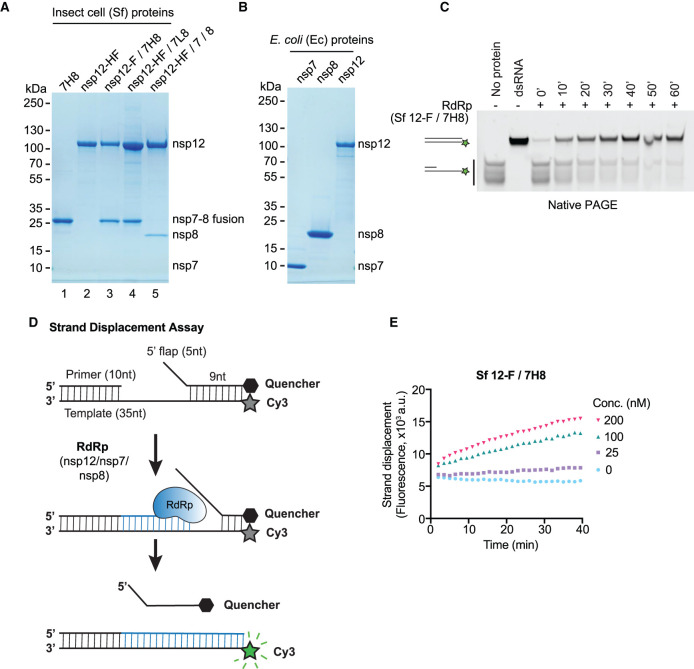
Figure 1. Development of a FRET-based SARS-CoV-2 RdRp strand displacement assay.
(A) Purified SARS-CoV-2 RdRp proteins expressed in baculovirus-infected insect cells (Spodoptera frugiperda, Sf) analysed by SDS–PAGE and Coomassie staining. 7H8: nsp7-His6-nsp8, nsp12-HF: nsp12-His6-3xFlag, nsp12-F/7H8: nsp12-3xFlag/nsp7-His6-nsp8, nsp12-HF/7L8: nsp12-His6-3xFlag/nsp7-GGSGGS-nsp8, nsp12-HF/7/8: nsp12-His6-3xFlag/nsp7/nsp8. (B) Bacterially expressed and purified SARS-CoV-2 nsp7, nsp8 and nsp12 proteins analysed by SDS–PAGE and Coomassie staining. The proteins were expressed as 14His-SUMO fusion proteins in E. coli. 14His-SUMO was removed by a SUMO-specific protease during purification generating native N-termini. (C) Gel-based primer-extension assay to test RNA-dependent RNA synthesis using the RdRp preparation Sf nsp12-F/7H8. The substrate consists of a 10 nt RNA primer annealed to the 3′ end of a 35 nt RNA template. The 5′ end of the template strand is labelled with a Cy3 fluorophore. Reaction products were analysed by native PAGE and visualisation of Cy3 fluorescence. Formation of duplex RNA by RdRp was observed over time. Controls: a preformed Cy3-labelled dsRNA with the same size as the reaction product (dsRNA), the primed substrate (no protein). (D) Schematic diagram illustrating the FRET-based RdRp strand displacement assay. The RNA substrate is composed of a Cy3 fluorophore-containing template strand, an annealed primer and an annealed quencher strand with a 5′ flap. RdRp activity synthesises RNA by extending the primer strand and displaces the quencher strand. The displaced quencher strand can no longer anneal to fully synthesised duplex RNA leading to an increase in Cy3 fluorescent signal. (E) FRET-based strand displacement assay using the indicated concentrations of Sf nsp12-F/7H8.