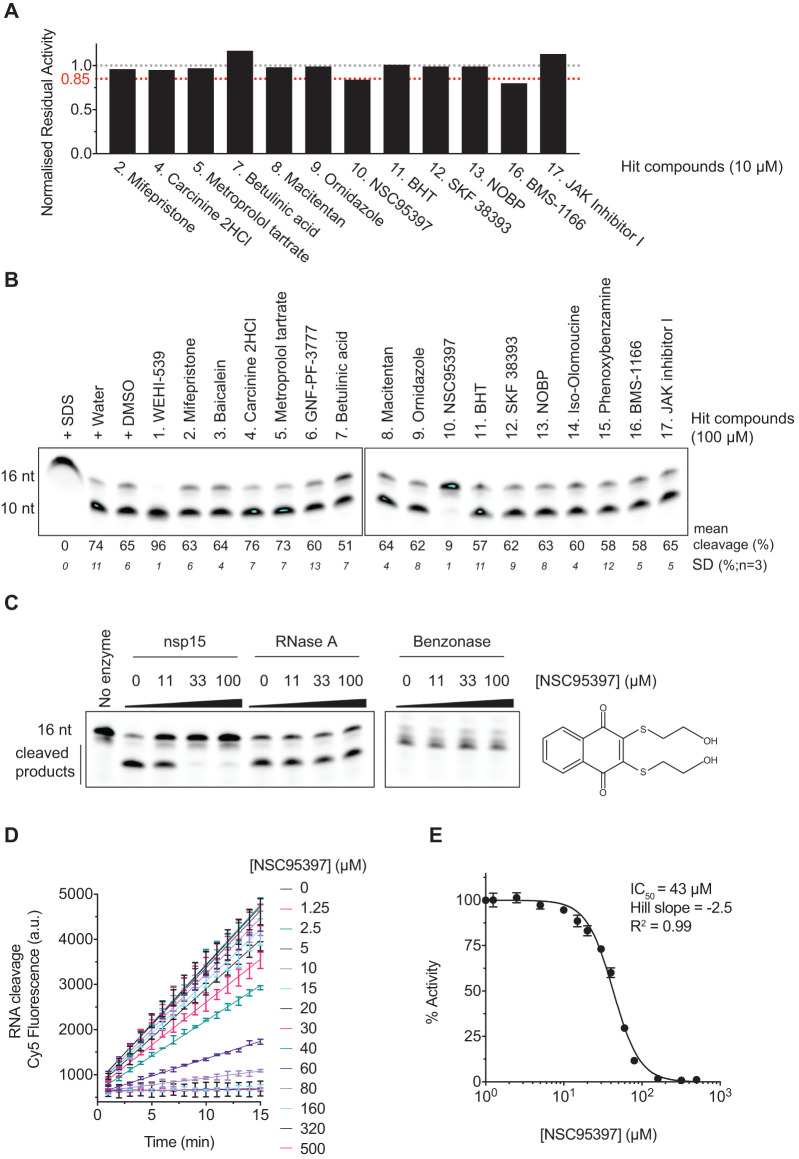
Figure 4. NSC95397 inhibits SARS-CoV-2 nsp15 endoribonuclease activity in vitro.
(A) Normalised residual activity of nuclease reactions monitoring cleavage of the 6 nt U substrate (500 nM) in solution using a Spark Multimode microplate reader (Tecan) in the presence of 75 nM of nsp15 and 10 µM of each of the 12 non-quenching selected screen hits (Supplementary Figure S3). Residual activities were calculated for each compound from experiment shown in Supplementary Figure S4, and normalised to control reaction. (B) Nuclease reactions containing 500 nM nsp15 enzyme and 1 µM 16 nt U substrate in the presence of 100 µM of each of the 17 selected screen hits. Reactions were performed for 20 min at 30°C and resolved in a denaturing TBE-urea denaturing polyacrylamide gel. Control lanes include enzyme denaturation by SDS and addition of water or DMSO, to mimic addition of drugs diluted either in water or DMSO. (C) Nuclease reactions containing 500 nM nsp15, 1 pg/µl RNase A or 25 mU/µl Pierce Universal Nuclease (benzonase) and 1 µM 16 nt U substrate in the presence of 0, 11, 33 or 100 µM NSC95397 inhibitor. Reactions were performed as in B. (D) Titration of NSC95397 inhibitor (0–100 µM) in the presence of 75 nM nsp15 enzyme and 500 nM 6 nt U substrate and fluorescence quantified in a Spark Multimode microplate reader (Tecan) at RT every minute for 15 min. (E) Dose-response curves and IC50 values of NSC95397 for SARS-CoV-2 nsp15. IC50 values were calculated as described in Experimental Procedures.