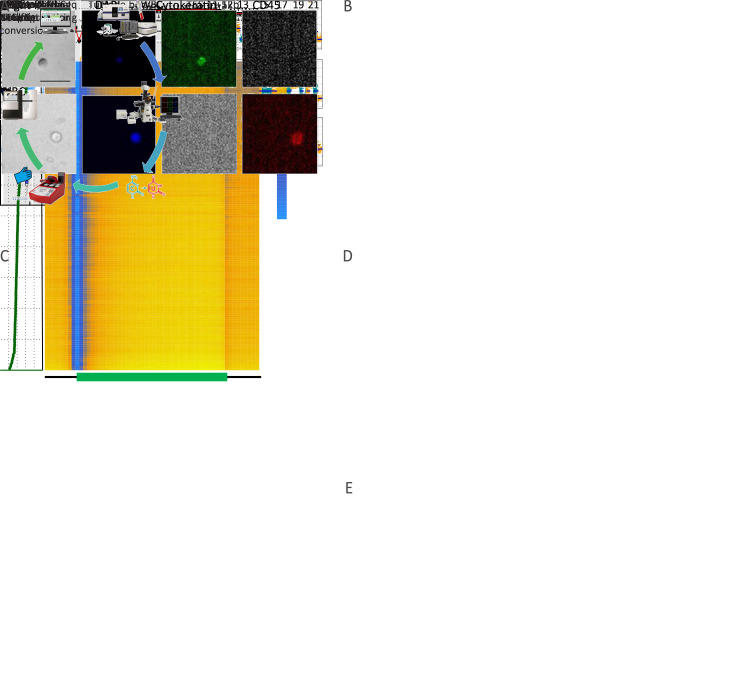
Figure 1.
Single-cell genome-wide bisulfite sequencing of cancer patient CTCs and tumor tissues. (A) Workflow illustrating an overview of the experimental steps. Standard blood samples obtained from cancer patients were used for CTC enrichment via the CellSearch® system, authentication from experience, and isolation by micromanipulation; (B) Representative immunofluorescence images of a CTC (top row) and a WBC (bottom row) co-stained with DAPI, anti-cytokeratin (CK), and anti-CD45. A CTC was identified as DAPI+, CK+, and CD45−, whereas a WBC was defined as DAPI+, CK−, and CD45+. Scale bar, 20 μm; (C) DNA methylation pattern in gene body regions as determined from all samples’ scBS-seq data. A total of 123 single cells and 73 nuclei were sequenced in the present study. The averaged DNA methylation level of CpG sites was calculated from all annotated RefSeq genes in gene body and their 5 kb flanking regions; (D) Box plot of the average methylation level of CTCs (blue), normal nuclei (orange), tumor nuclei (green), and WBCs (red); (E) CNA profiles from Patient SC6 (shown as an example) at 500 kb resolution. The top 3 panels show the CNA pattern of CTCs, whereas the bottom panel shows the normal copy number pattern of the WBC control. CTC, circulating tumor cell; WBC, white blood cell; DAPI, 4’,6-diamidino-2-phenylindole; TSS, transcription start site; TES, transcription end site. ***, P<0.001.