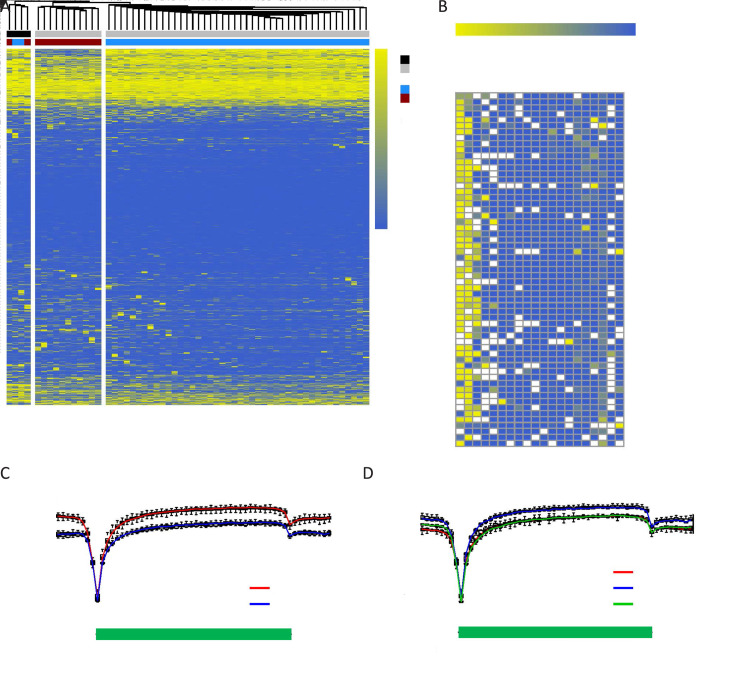
Figure 2.
Inter- and intra-patient epigenetic heterogeneity of CTCs in SCLC. (A) Unsupervised hierarchical clustering of the methylome of individual CTCs (grey) and WBCs (black) derived from Patient SC6 (blue) and Patient SC7 (red). All RefSeq gene promoters were used for the analysis. The color key from blue to yellow indicates low to high methylation level; (B) DNA methylation heatmap of individual CTCs and WBCs derived from Patients SC6 and SC7. A total of 20 known SCLC-associated gene promoters were used for the analysis. The color key from blue to yellow indicates low to high methylation level, whereas white checks indicate unavailable data; (C,D) DNA methylation pattern in gene body regions of CTCs from Patients SC6 and SC7 with SCLC (C), and Patients ADC1, ADC3, and ADC6 with ADC (D). The averaged DNA methylation level of CpG sites was calculated from all annotated RefSeq genes in gene body and their 5 kb flanking regions. The line represents the mean value for CTCs from each patient: Patient SC6 (left, red), Patient SC7 (left, blue), Patient ADC1 (right, red), Patient ADC3 (right, blue), and Patient ADC6 (right, green). CTC, circulating tumor cell; SCLC, small-cell lung cancer; ADC, adenocarcinoma; TSS, transcription start site; TES, transcription end site.