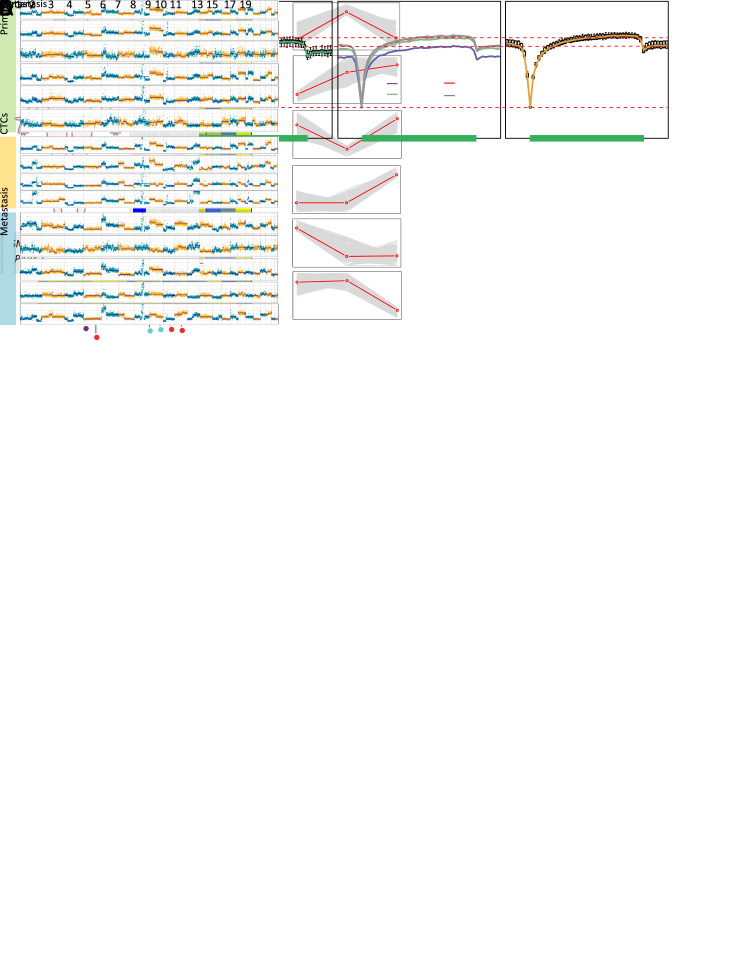
Figure 4.
Different evolutionary histories traced from copy number and methylation profiles in Patient GC1. (A) Left: a diagram of tumor progression in Patient GC1 who was diagnosed with GC accompanied by abdominal ovarian metastasis; right: CNA deduction results of scBS-seq datasets from Patient GC1. The copy numbers (blue and red dots) were plotted along the genome at a bin size of 500 kb. The ordinate coordinate represents copy numbers ranging from 0 to 6 (a copy number >6 was set to 6); (B) CNA regions containing known GC-associated genes. Deletions (blue) and amplifications (red) are shown; (C) DNA methylation in gene body regions of normal nuclei (purple), primary tumor nuclei (green), CTCs (multi-colored), and metastatic tumor nuclei (orange) derived from Patient GC1. Average CpG methylation levels along the scaled gene bodies, 5 kb upstream of TSS, and 5 kb downstream of TES for all RefSeq genes were used for the analysis; (D) Methylation heatmap of DMRs among different samples from Patient GC1 (blue, individual cells; red, pseudo-bulk sample merged with individual cells of the same cell type). Cell types were color coded: green (primary tumor nuclei), purple (CTCs), and orange (metastatic tumor nuclei). The color key from blue to yellow indicates low to high methylation level. The line charts show representative DNA methylation patterns during tumor metastasis; (E) Phylogenetic (top) and phyloepigenetic (bottom) reconstruction showing evolutionary histories in Patient GC1 inferred from the chromosomal breakpoint profiles (top) and DNA methylation distance matrix (bottom). Cell types were color coded: peacock green (primary normal cells), red (CTCs), purple (primary tumor cells), and turquoise (metastatic tumor cells). The Hamming distance metrics are individual and non-comparable. GC, gastric cancer; CNA, copy number alteration; CTC, circulating tumor cell; TSS, transcription start site; TES, transcription end site; DMR, differentially methylated region.