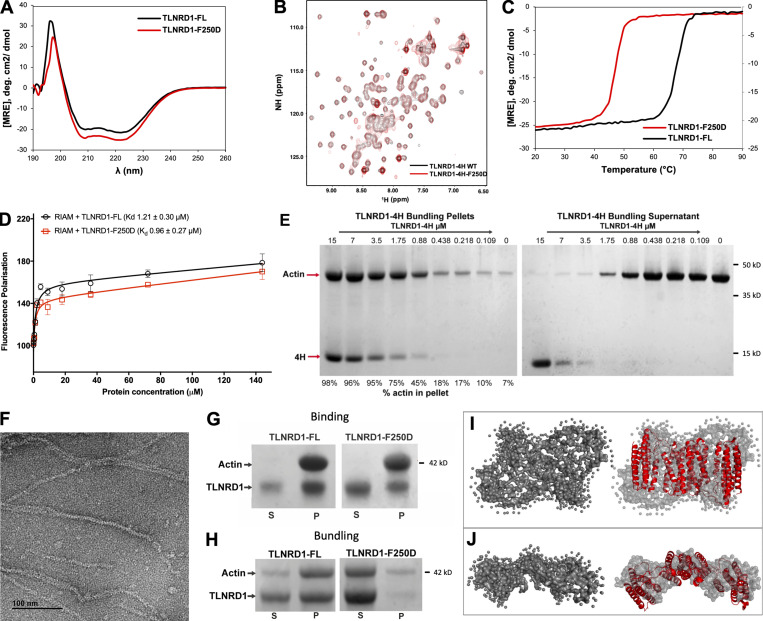
Figure S2.
Biochemical analysis of TLNRD1. (A) CD spectra of TLNRD1-FL (black) and TLNRD1-F250D (red) showing no change in overall secondary structure as a result of the mutation. (B) nNMR analysis of the F250D mutant shows it closely resembles the WT. 1H,15N heteronuclear single quantum coherence (HSQC) spectra of 150 µM 15N-labeled TLNRD1–4H WT (black) or TLNRD1-4H-F250D (red). (C) CD thermostability analysis of TLNRD1-FL and TLNRD1-F250D showing a reduction in stability with loss of dimerization. (D) Binding of fluorescein-labeled RIAM (residues 4–30) peptide with TLNRD1-FL WT (black) and TLNRD1-FL F250D (red) measured using a fluorescence polarization assay. (E) Low-speed actin cosedimentation bundling assay with TLNRD1-4H serial dilution against 15 µM F-actin. The two gels show the (left) pellet and (right) supernatant fractions. At high TLNRD1-4H concentrations, unbound TLNRD1-4H is present in the supernatant, suggesting the binding becomes saturated. At low TLNRD1-4H concentrations, the actin is predominantly in the supernatant. (F) Actin filaments alone visualized by negative stain EM. (G and H) Actin binding of the F250D mutant. (G) High-speed actin binding assay of TLNRD1-FL and TLNRD1-F250D showing little impact on actin binding with loss of dimerization. (H) Low-speed actin cosedimentation assay showing loss of TLNRD1 actin bundling with loss of dimerization as a result of the F250D mutation. (I and J) SAXS analysis of TLNRD1 shows compact domain arrangement in solution. (I) SAXS envelope reconstruction with GASBOR, showing the best fit with the TLNRD1-FL crystal structure. (J) Top-down view of I. MRE, molar residue ellipticity; deg., degrees; NH, amide.