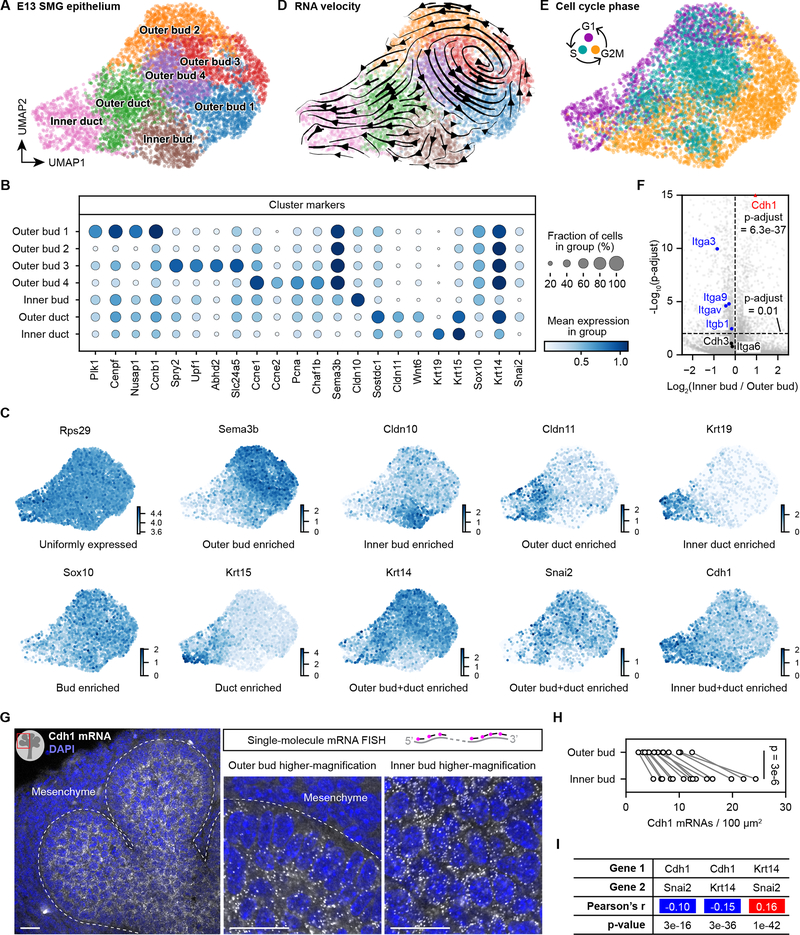
Figure 4. Single-cell transcriptome profiling reveals spatial transcriptional patterns of the branching salivary gland epithelium.
(A, D, E) Scatter plots of 6,943 single-cell transcriptomes from the E13 mouse salivary gland epithelium shown in UMAP embedding and color coded by clusters (A and D) or cell cycle phase (E). Each dot represents one cell. Arrows in (D) indicate local RNA velocity estimated from unspliced and spliced transcripts of nearby cells. (B) Dot plot of selected cluster marker genes. (C) Scatter plots of single-cell transcriptomes of E13 salivary gland epithelium in UMAP embedding and color coded by the expression level of indicated genes. (F) Volcano plot comparing the expression levels of integrin and cadherin genes in the Inner bud vs. Outer bud (combining Outer bud 1–4). p-adjust, t-test with Benjamini-Hochberg correction. (G) Confocal images of Cdh1 mRNAs detected by single-molecule mRNA FISH in an E13 salivary gland. Each white dot is one Cdh1 mRNA molecule. (H) Plot of the Cdh1 mRNA density in outer or inner epithelial bud of E13 salivary glands. Measurements from the same image were connected by a line. p-value, paired two-sided t-test. (I) Table of the Pearson’s correlation coefficients and p-values between indicated genes. Blue and red shadings indicate negative and positive correlations, respectively. Scale bars, 20 μm. See also Figure S4.