Figure 1. DINO, a lncRNA gene recurrently methylated in human cancers, is a haplo-insufficient tumor suppressor.
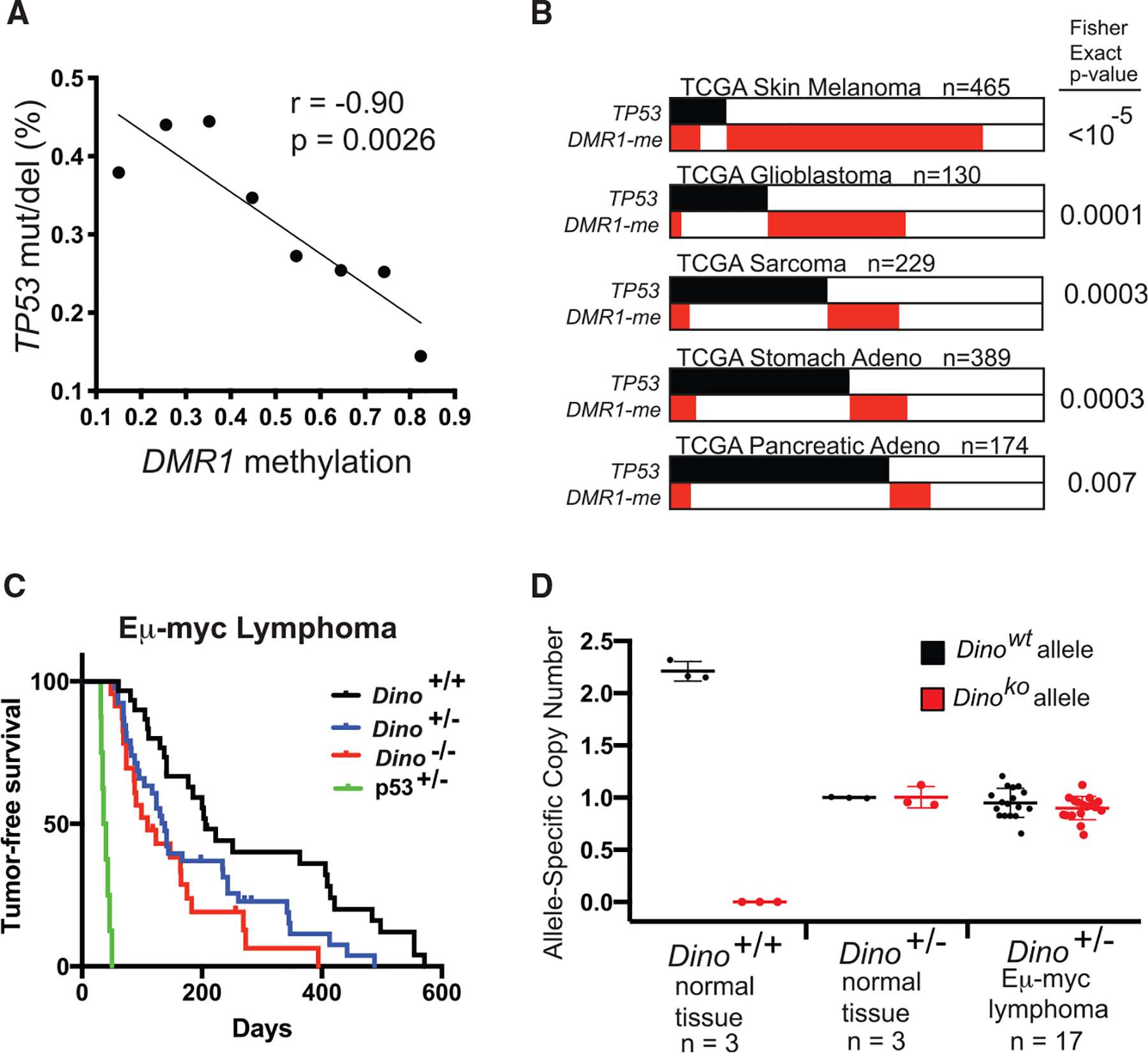
(A) Rate of TP53 somatic SNVs or deletions in 8,141 samples from the TCGA Pan-Cancer dataset in relation to methylation of DMR1. Samples are binned according to average DMR1 methylation in increments of 0.1 from <0.2 to ≥0.8, p value for correlation for 8 graphed data points.
(B) Mutual exclusivity analysis of DINO methylation (average methylation of DMR1 ≥0.5) and TP53 somatic SNVs or deletions in indicated TCGA datasets, Fisher exact test.
(C) Lymphoma-free survival of Eμ-myc mice of indicated genotype. Median lymphoma-free survival in Dino−/− mice occurs at 109 days (n = 23, p = 0.0007, log-rank test) and 138 days in Dino+/− mice (n = 40, p = 0.01, log-rank test) compared to 206 days in Dino+/+ mice (n = 30). Survival of p53+/− Eμ-myc mice are shown for comparison (median lymphoma-free survival 38 days, n = 8).
(D) Dino allele copy number in Eμ-myc Dino+/− lymphomas compared to indicated normal tissue controls as quantified by qPCR, mean ± SD.
See also Figures S1 and S2 and Tables S1, S2, and S3.
