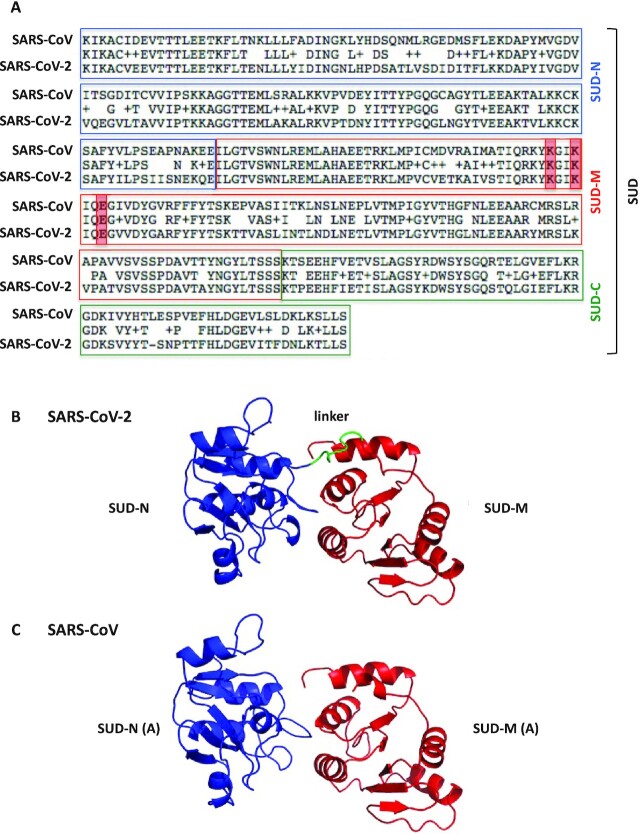
Figure 1.
Primary structure alignment of the SARS-CoV and SARS-CoV-2 SUD proteins and homology modeling of the SARS-CoV-2 SUD-NM protein. (A) The alignment was performed using the BLAST algorithm with the Blosum62 scoring matrix between amino acids 389–720 and 413–744 of SARS-CoV (NC004718) and SARS-CoV-2 (BetaCoV_Wuhan_IVDC-HB-01_2019) Nsp3 proteins, respectively. More precisely, the SUD-N and SUD-M macrodomains and the SUD-C frataxin-like domain of these two viruses share 88.2, 96.1 and 91.4% amino-acid sequence similarities (ExPASy LALIGN algorithm). The K565-K568-E571 residues (highlighted in red), present in SARS-CoV SUD-M and important for viral replication (5) are conserved in SARS-CoV-2 SUD-M. (B) Ribbon representation of the modeled structure of the SARS-CoV-2 SUD-NM computed with the Modeller software using the SARS-CoV SUD-NM protein as template structure (PDB code: 2W2G (8)). SUD-N is in blue, SUD-M in red and the 518–524 linker connecting the two macrodomains is in green. (C) Structure of the SARS-CoV SUD-NM monomer A template structure (PDB code: 2W2G) provided for comparison and in the same orientation than in (B).