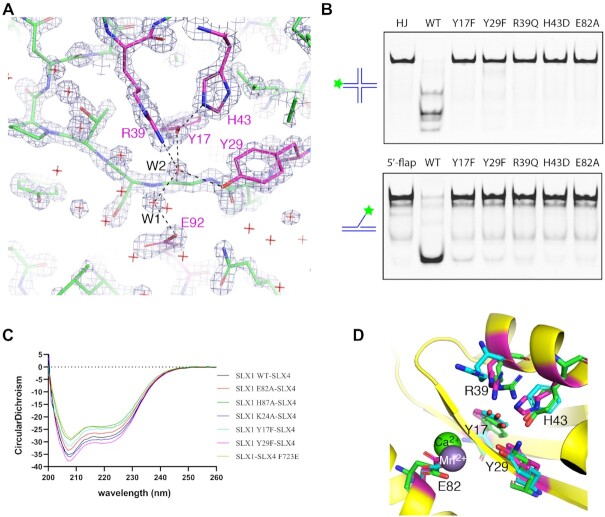
Figure 2.
The catalytic active site of SLX1. (A) A section of the 2Fo – Fc electron density map, contoured at 3.5 σ, covering the active site. A stick model of the refined structure is superimposed. The carbon half-bonds of the active site residue are labeled magenta, and the rest is shown in green, while nitrogen and oxygen half-bonds are shown in blue and red, respectively. Red crosses indicate water molecules, and two of which making hydrogen bonds to catalytic residues are designated W1 and W2. (B) Mutation of SLX1 active site residues results in impaired cleavage of HJ (top panel) and 5′-flap DNA (bottom panel) substrates. The assay was carried out using the protein complexes assembled from wild-type (WT) or indicated SLX1 mutants assembled with SLX4SAP+CCD, and a 5′-Cy3 labeled HJ and 5′-flap DNA as the substrates (Supplementary Table S1). (C) Circular dichroism (CD) spectra of WT and mutant SLX1–SLX4SAP+CCD complexes show that protein folding was not compromised by mutations. (D) Superposition of Hpy188I (green; PDB ID: 3OR3) and UvrC (cyan; PDB ID: 1YD0) GIY-YIG domains with the Uri/GIY-YIG domain of SLX1 shows that the active sites are highly conserved. A calcium ion from the Hpy188I structure (green sphere), and a manganese ion (grey sphere) in the UvrC structure offer insights into the role of the conserved glutamate residue.