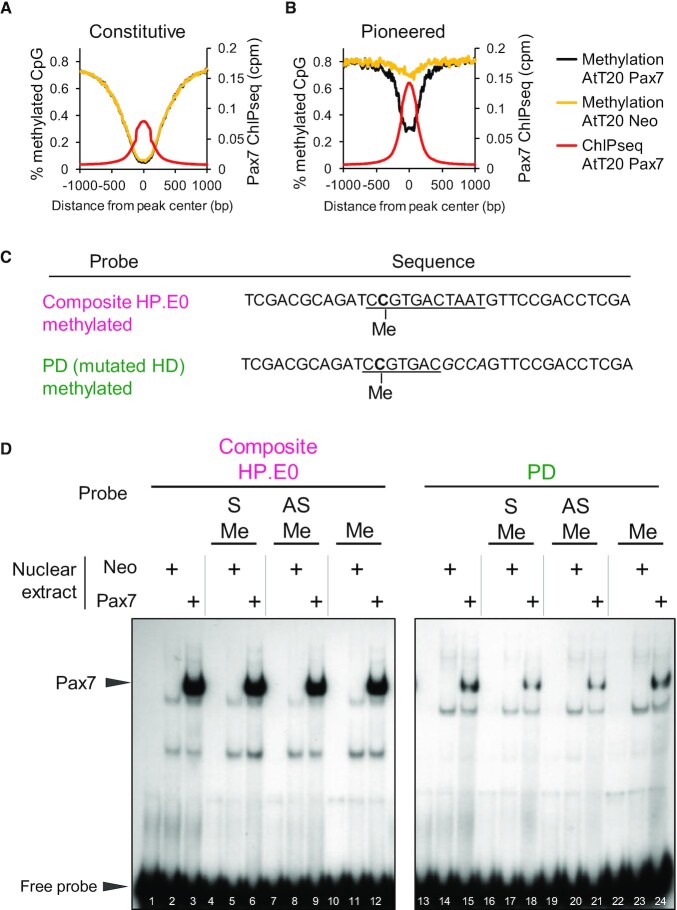
Figure 3.
Pax7 binding is independent of DNA methylation. (A, B) Average profiles of DNA methylation (% methylated CpG) around Pax7 ChIPseq peaks present at Constitutive (A) and Pioneered (B) enhancers. Data are shown for control AtT20-neo compared to AtT20-Pax7 cells and aligned with the average Pax7 ChIPseq profiles. (A) DNA sequences of probes used in EMSA to test the effect of DNA methylation on Pax7 binding. Methylated cytosine (C) residues are shown on sense (S) strand. (B) In vitro binding (EMSA) of Pax7 to either composite or PD probes methylated or not on sense (S), antisense (AS) or both (Me) strands.