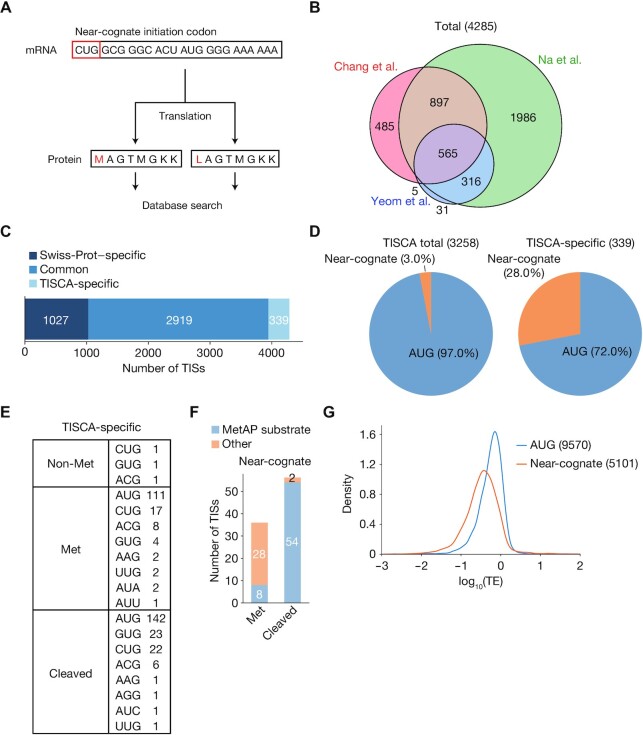
Figure 4.
Proteomics analysis of TISs identified by TISCA. (A) Outline for the generation of amino acid sequence lists for comparison with proteomics data. For the ORFs with near-cognate initiation codons identified by TISCA, two types of sequences were generated: one in which the first amino acid was the residue actually encoded by the corresponding codon, and the other in which the first amino acid was methionine regardless of the encoded residue. (B) Venn diagram showing numbers of TISs identified in the three data sets generated by the proteomics analyses of Chang et al. (37), Na et al. (19) and Yeom et al. (36). (C) Classification of TISs identified by proteomics analyses. The TISs were classified as those identified only in Swiss-Prot, both in Swiss-Prot and by TISCA, or by TISCA alone. (D) Pie charts indicating initiation codon usage of TISs identified from all TISCA data or from TISCA-specific data. (E) Number of novel TISs sorted by the type of initial amino acid and initiation codon. (F) Number of novel TISs in the Met or cleaved categories that were identified as possible substrates for MetAP. ORFs containing alanine, cysteine, glycine, proline, serine, threonine, or valine as the second amino acid were considered to encode potential MetAP substrates. (G) Density plots of translation efficiency (TE) as determined by Ribo-seq analysis for ORFs initiated at AUG or near-cognate codons. Multiple overlapping ORFs were excluded from the analysis.