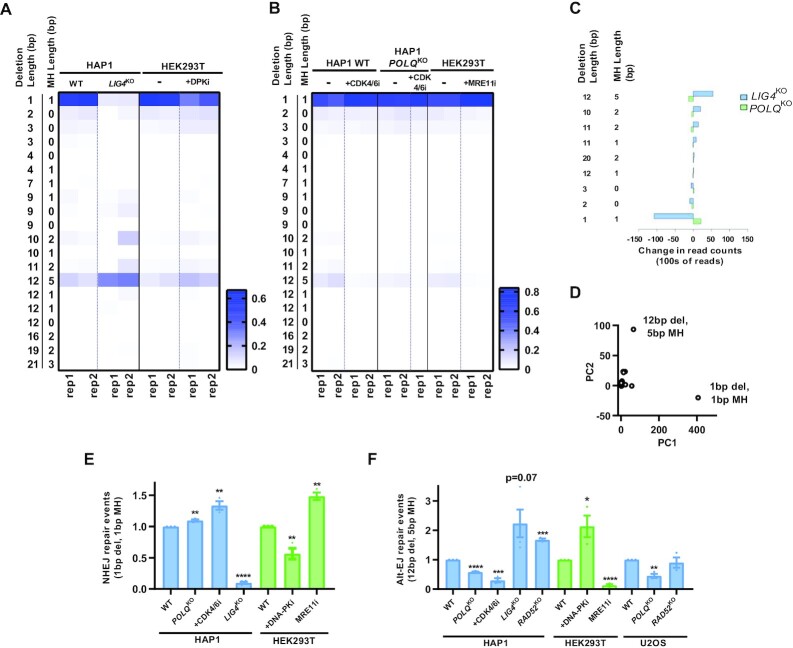
Figure 3.
Single characteristic reads for each pathway represent most Alt-EJ and NHEJ-specific information. (A) Heat map of the most frequent 20 reads containing a deletion or insertion. In wild type cells, a 1 bp deletion is most common which is also heavily suppressed in a LIG4 knockout context or with pharmacologic inhibition of DNA-PK. A 12 bp deletion with 5 bp of surrounding microhomology is induced in these same contexts. (B) The 12 bp deletion with 5 bp of microhomology is dependent upon polymerase theta (POLQ) and cell cycle dependent (CDK4/6 inhibition) and relies on MRE11 implying a resection-mediated process. (C) Specific reads which depend upon LIG4 or POLQ. (D) Principal component analysis of reads most discriminating between NHEJ and Alt-EJ. (E-F) Quantification of the 1bp and 12 bp/5 bp MH read normalized to the WT condition. Mean ± SEM from 3–4 experiments shown. Asterisks signify t-tests as follows: * P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001.