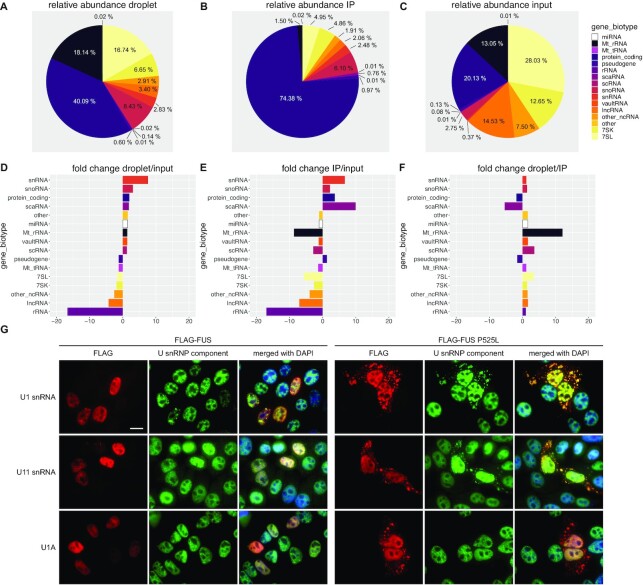
Figure 3.
RNA interactome of LLPS and non-LLPS FUS. (A) Relative abundance of different RNA species co-purified with LLPS FUS (FUS droplets). (B) Relative abundance of different RNA species co-purified with non-LLPS FUS (co-IP). (C) Relative abundance of different RNA species in the input sample. (D) fold change of relative RNA abundance of different RNA species comparing RNAs found under LLPS conditions relative to the input. (E) Same as in D, but comparing RNA found in non-LLPS conditions compared to the input. (F) Same as in D, but comparing RNA abundance between LLPS and non-LLPS conditions. (G) HeLa cells transiently transfected with either FLAG-FUS (left) or FLAG-FUS P525L (right) and stained for FLAG (red channel) and different components of the U1 snRNP (green channel), either by RNA-FISH (U1 and U11, first and second row) or immunostaining (U1A, third row). Cells were counterstained using DAPI. Scale bar = 15 μm.