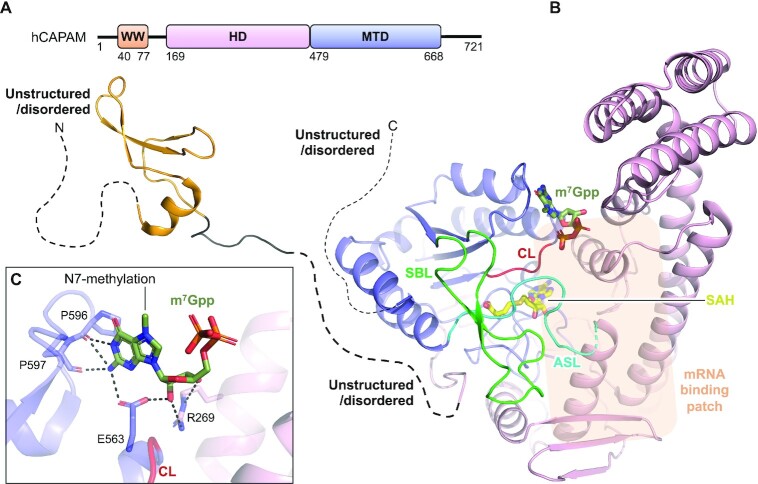
Figure 4.
Structure of human CAPAM. (A) Schematic outline of the human CAPAM (hCAPAM) protein domains. Crystallized parts are indicated in boxes. (B) Structure of human CAPAM with the WW (orange) (PDB 2JX8), the SAH-bound (yellow) MTD (blue), and the HD (pink) (PDB 6IRW). The active site loop (ASL, cyan) that connects the MTD and HD is partly disordered in human CAPAM and is therefore modelled based on zebrafish CAPAM (PDB 6IRX). m7Gpp (dark green) is modelled from zebrafish CAPAM (PDB 6IS0). The catalytic loop (CL, red) and substrate binding loop (SBL, green) are shown. The mRNA substrate binding patch is indicated with a transparent orange box. The sections not included in the structures are shows as dotted lines, and their structure/order state is indicated. (C) Zoom of the active site showing the build part (m7Gppp) of the m7Gpp-derived ligand, co-crystallized with zebrafish CAPAM (PDB 6IS0). The enzyme residues that interact with the ligand are shown in sticks and labelled. Hydrogen bonds are shown in black, dashed lines, none of which are with the N7-methylation.