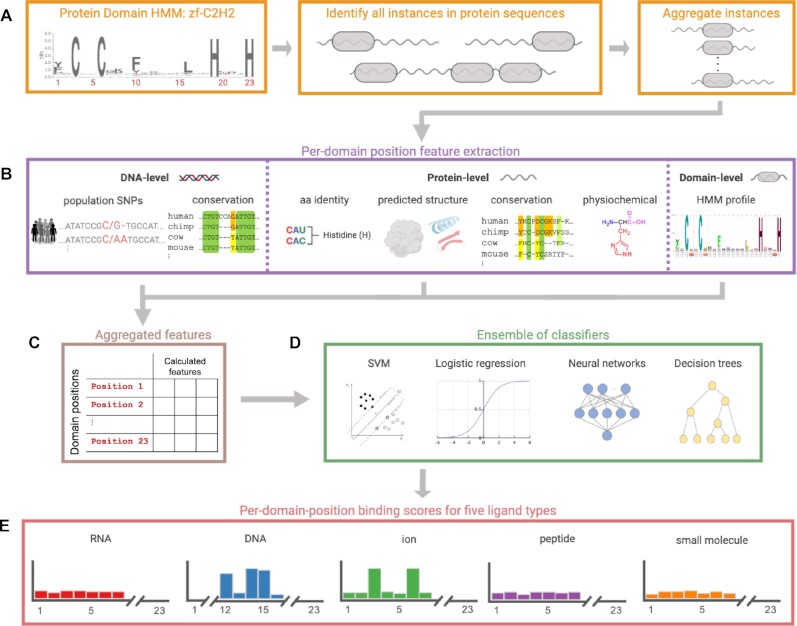
Figure 1.
Workflow for predicting binding positions in protein domains. (A) For a given protein domain (represented as an HMM), instances of the domain are found across all human proteins and aggregated to construct per-position features. The zf-C2H2 domain (PF00096) is shown as an example. (B) For instances of the domain, features for each position are calculated at either the DNA base, protein amino acid or whole-domain level. The figure illustrates a few example features calculated at each level. DNA-level features (left) include population allele frequencies and evolutionary conservation. Protein amino acid-level features (middle) include amino acid identity, information derived from predicted structure (e.g., secondary structure and solvent accessibility), amino acid conservation across orthologs, and physicochemical properties of the amino acids. Domain-level features (right) include the HMM emission probabilities and the predominant amino acid at each position. (C) Features from the three levels are aggregated across instances for each protein domain position. (D) Using these features and a set of known DNA, RNA, ion, peptide and small molecule binding positions within domains, we train a heterogeneous ensemble of classifiers to identify positions binding each ligand type. (E) Results from these classifiers are combined and each final model outputs per-domain-position binding scores for one of the ligand types (Figure generated using biorender.com).