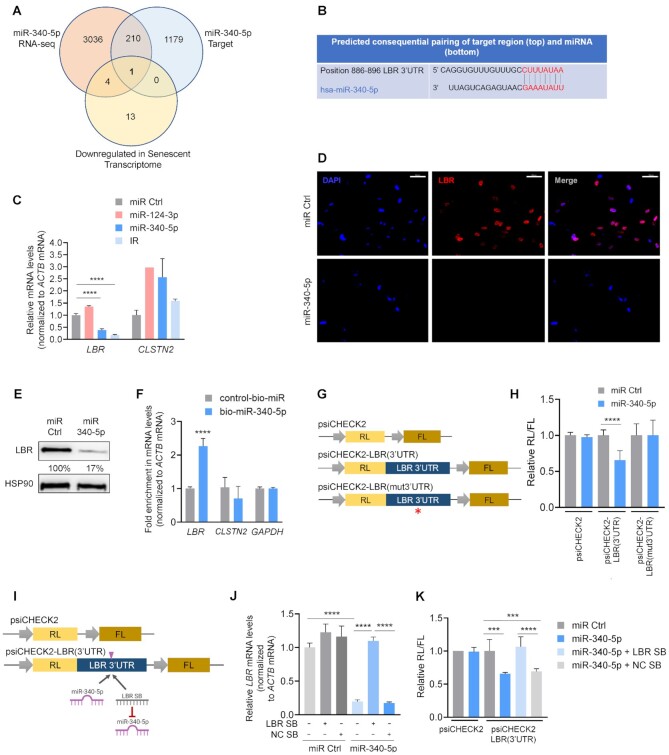
Figure 4.
LBR mRNA is a direct target of repression by miR-340-5p. (A) Venn diagram comparing RNAs reduced by miR-340-5p overexpression as determined by RNA-seq analysis, predicted miR-340-5p-target transcripts and transcripts decreased in the senescent transcriptome as identified by RNA-seq analysis. (B) Alignment of miR-340-5p with the LBR 3′UTR seed sequence as predicted by TargetScan. (C) RT-qPCR analysis of the levels of LBR and CLSTN2 mRNAs in WI-38 fibroblasts (PDL20) 4 days after transfection with miR Ctrl, miR-124-3p, miR-340-5p or 10 days after IR. mRNA levels were normalized to ACTB mRNA levels. (D) Immunofluorescence micrographs (40×) to detect nuclei (DAPI, blue) and LBR (red) in WI-38 fibroblasts (PDL20) 4 days after transfecting miRNA Ctrl or pre-miR-340-5p. Scale bar: 50 μm. (E) Western blot analysis of the levels of LBR or loading control HSP90 in WI-38 cells processed as explained in panel (D). The % values represent the densitometric calculations of the signals on the western blots, shown as a percentage relative to miR Ctrl and normalized to HSP90 signals. (F) RT-qPCR analysis of the enrichment of LBR mRNA (as well as controls CLSTN2 mRNA and GAPDH mRNA) in the material pulled down with streptavidin-conjugated magnetic beads 16 h after transfecting cells with either biotinylated miR-340-5p (bio-miR-340-5p) or a control biotinylated RNA (control-bio-miR). (G) Luciferase reporter constructs. The dual reporter vector psiCHECK2 expresses Renilla luciferase (RL) and firefly luciferase (FL) under separate promoters. Plasmid psiCHECK2 is the empty vector control; plasmid psiCHECK2-LBR(3′UTR) has the LBR 3′UTR inserted directly following the RL coding region; plasmid psiCHECK2-LBR(mut3′UTR) has the LBR 3′UTR with a mutation in the miR-340-5p binding site (red asterisk) inserted directly following the RL coding region. (H) Proliferating WI-38 fibroblasts were transfected with psiCHECK2, psiCHECK2-LBR(3′UTR) or psiCHECK2-LBR(mut3′UTR) plasmids; 24 h later, cells were transfected with miR Ctrl or miR-340-5p, and 24 h after that, lysates were prepared and RL activity was normalized to FL activity and further normalized to miR Ctrl transfection for each plasmid. (I) Luciferase reporter constructs depicting the position where miR-340-5p and the LBR SB bind is indicated (pink arrowhead). (J) RT-qPCR analysis of LBR mRNA levels 4 days after co-transfection of either miR Ctrl alone or miR-340-5p in the presence or absence of a negative control site blocker (NC SB) or a site blocker specifically designed to abrogate binding of miR-340-5p to the LBR mRNA (LBR SB). (K) Assessment of LBR 3′UTR activity. Proliferating WI-38 fibroblasts were transfected with psiCHECK2 or psiCHECK2-LBR(3′UTR) plasmids; 24 h later, cells were transfected with miR Ctrl, miR-340-5p, miR-340-5p + LBR site blocker (LBR SB) or miR-340-5p with a negative control site blocker (NC SB). RL activity was normalized to FL activity and further normalized to miR Ctrl transfection for each plasmid. Data in panels (C), (F), (H), (J) and (K) represent the mean values ± SD from three biological replicates. Significance was established using Student’s t-test. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.