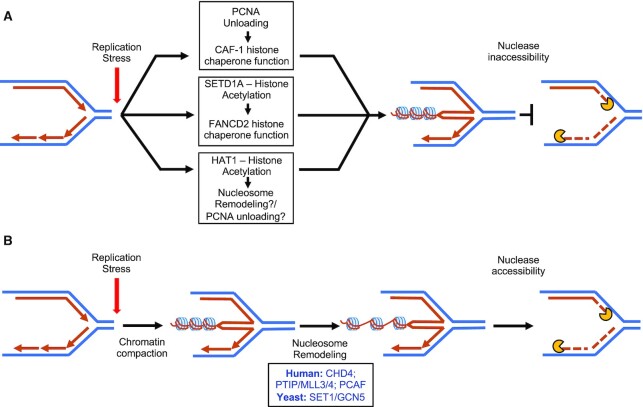
Figure 2.
The impact of histone modifications and nucleosome remodeling on replication fork stability. (A) The ability to mediate efficient histone chaperone activity by CAF-1 and FANCD2 is associated with replication fork protection. The chaperone activity of CAF-1 is enabled by efficient PCNA-unloading from the lagging strand, which in turn depends on Okazaki fragment ligation and efficient lagging strand gap-filling by ubiquitinated PCNA. The fork protective histone chaperone activity of FANCD2 depends on the histone methyltransferase activity of SETD1A, which in turn restricts the activity of the CHD4, a member of the repressive NuRD complex. HAT1 promotes replication fork protection putatively through histone acetylation as well as ensuring timely PCNA unloading. (B) Nucleosome remodeling may contribute to fork degradation by enhancing the activity of nucleases to reversed fork substrates. CHD4 enables the resection of stalled replication forks in cells deficient in BRCA2 and FANCD2, by promoting the activity of MRE11. The methyltransferase activity of the PTIP interacting histone methyltransferases MLL3/4 promote fork resection in BRCA-deficient cells by enabling the recruitment of MRE11. Similarly, the H4K8 acetyltransferase activity of PCAF promotes the recruitment of MRE11/EXO1 to stalled forks in BRCA-deficient cells. Finally, in yeast models, the methyltransferase and acetyltransferase activities of SET1 and GCN5 respectively are required for nucleosome remodeling events enabling the MRX complex to resect DNA at stalled replication forks.