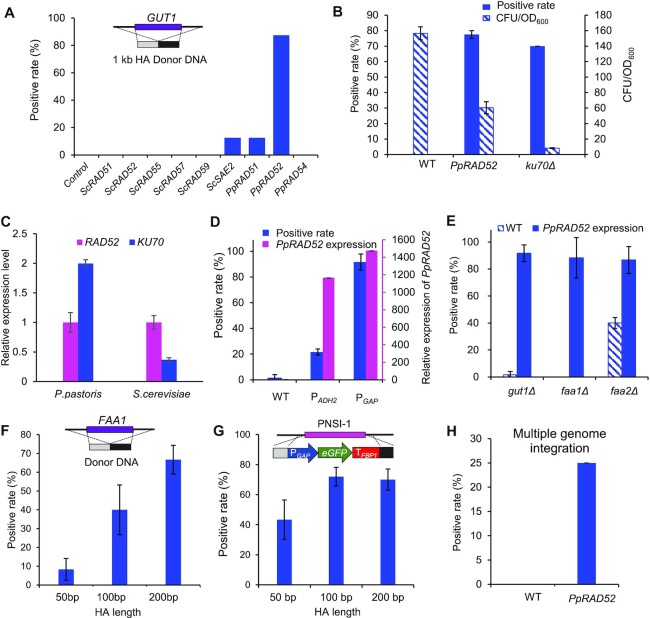
Figure 2.
Enhancing homologous recombination (HR) for precise genome editing. (A) Screening HR related genes for improving seamless deletion. The heterologous HR related genes from S. cerevisiae or endogenous P. pastoris genes were overexpressed and be followed by deleting GUT1 in P. pastoris. The strains carried the plasmid expressing Cas9 and gRNA for targeting GUT1. The deletion cassette was constructed by fusing the upstream and downstream homologous arms of 1 kb. Wild-type strain GS115 without overexpressing RAD genes was used as a control. Eight colonies were picked for colony PCR analysis with primer pair Check-GUT1-F/Check-GUT1-R. (B) PpRAD52 overexpression improved the seamless deletion of FAA1 compared with KU70 deletion. (C) Relative expression level of KU70 and RAD52 in S. cerevisiae or P. pastoris. The housekeeping actin gene was chosen as baseline for quantification of expression level. Relative expression level of these genes was calculated by normalized the expression level of RAD52 in the respective strains. (D) PpRAD52 overexpression improved the seamless deletion of GUT1 in P. pastoris. The PGAP and PADH2 were used to overexpress RAD52 and the relative expression levels of RAD52 were analyzed by qPCR. (E) PpRAD52 overexpression improved the seamless deletion of GUT1, FAA1, and FAA2. (F) HR rates for FAA1 deletion with various HA lengths in strains of PpRAD52 overexpression. (G) HR rates for eGFP overexpression cassette integration into PNSI-1 site with various HA lengths in strains of PpRAD52 overexpression. (H) The positive rates of integration of three genes at three genome loci. Twenty colonies were picked for colony PCR analysis. 1 kb homologous arms was used in both integration and knockout experiments unless otherwise noted. Mean values and standard deviations of biological triplicates were shown in 2B to 2H.