Figure 1. Isolation and identification of TNAP as a cold-inducible PCr’ase from mitochondria of thermogenic fat.
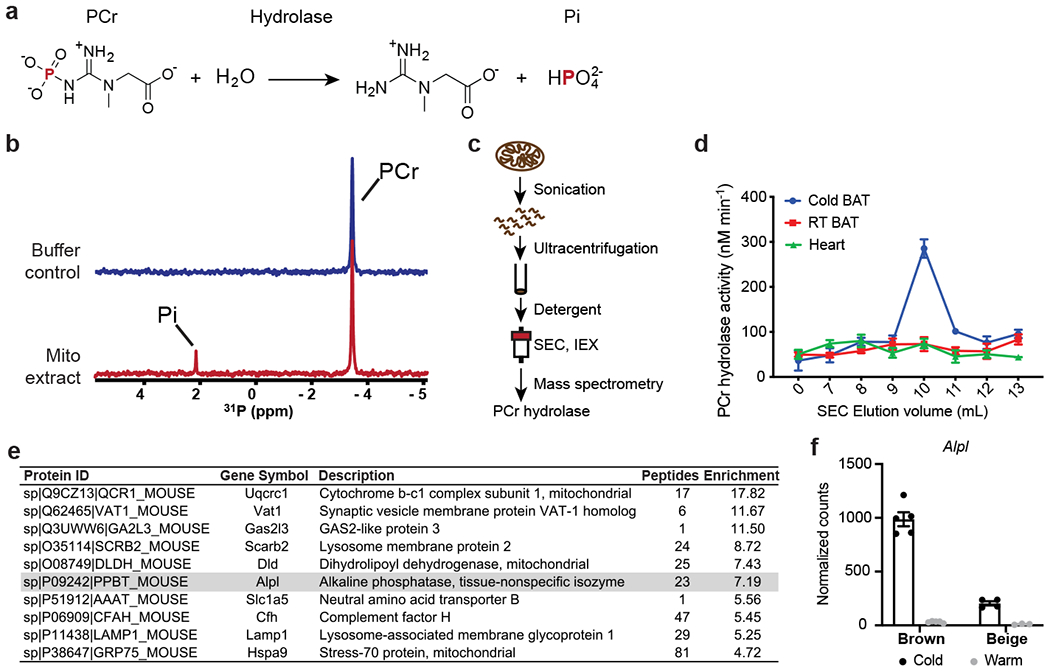
(a) Chemical equation for hydrolysis reaction of phosphocreatine, with phosphorus-containing chemical species indicated on top. PCr: phosphocreatine; and Pi: inorganic phosphate.
(b) Stacked 1-dimensional traces of 31P NMR frequency-domain spectra showing the occurrence of hydrolysis reaction of phosphocreatine with addition of mitochondrial protein extract (1.0 mg/ml) from brown adipose tissue (BAT) of cold-acclimated mice in 6 hours. All activities shown in rest of the figure were measured by 31P NMR, if not otherwise indicated.
(c) Scheme of isolation and identification of PCr phosphatase(s) from sucrose-gradient purified mitochondria preparation from murine tissues.
(d) PCr phosphatase activities of different fractions of size-exclusion chromatography. Error bars represent the standard errors of non-linear regression that fits a straight-line model to the initial linear phase of PCr hydrolysis kinetics measured by 31P NMR over 4 time points (shown in Source Data).
(e) List of top 10 proteins enriched in the active fraction versus a nearby less active fraction of IEX. TNAP (gene name: Alpl) is highlighted in grey.
(f) UCP1-TRAP (Translating Ribosome Affinity Purification) data showing the cold-sensitivity of Alpl expression in brown and beige adipocytes. Data were adapted from Roh, H. C. et al.23 and normalized with DESeq241; and are presented as means ± SEM; n=5 for cold and warm brown, 4 for cold beige, and 3 for warm beige (all are biologically independent samples).
