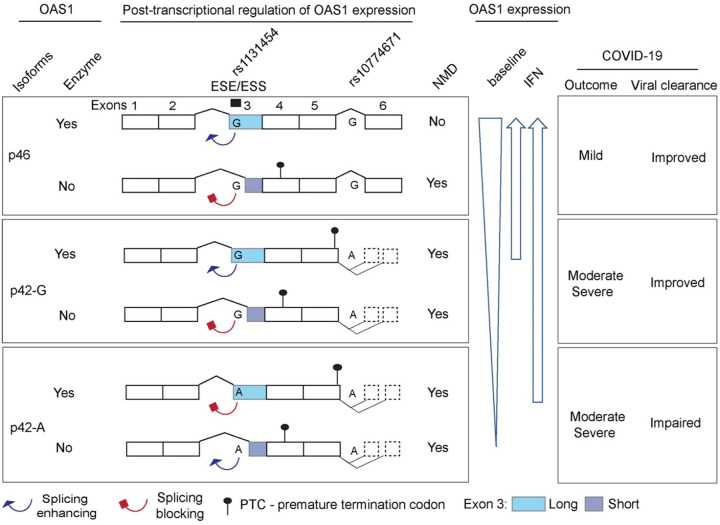
Figure 8. The proposed model for mechanisms underlying association between OAS1 genetic variants and COVID-19 outcomes.
Two OAS1 variants – exon 3 missense variant (rs1131454-A/G, Gly162Ser) and splice site variant (rs10774671-A/G) determine the structure and expression levels of OAS1 isoforms. Alleles of the splicing variant rs10774671 define the OAS1 isoforms – OAS1-p42 (A allele) and OAS1-p46 (G allele). Alleles of rs1131454 create an exonic splicing enhancer/silencer (ESE/ESS) for splicing of the canonical/Long vs. alternative/Short exon 3. Transcripts with Short exon 3 are terminated by premature stop codons (PTCs) within exon 4 and efficiently targeted by nonsense-mediated decay (NMD). The stop codon for OAS1-p42 is located within exon 5, followed by several additional exons creating OAS1-p44 and OAS1-p48 isoforms, making this stop codon a PTC. Thus, OAS1-p42 is also targeted by NMD, albeit less efficiently compared to transcripts with PTCs in exon 4. The combined splicing effects of rs10774671, which creates alternative OAS1 isoforms and rs1131454, which regulates the inclusion of Short or Long exon 3 and thus the introduction of additional PTCs result in variable degradation of OAS1 transcripts by NMD. At baseline, the expression is highest for OAS1-p46 and the lowest for OAS1-p42-A. However, treatment with IFNs may compensate for NMD, allowing OAS1-p42 protein to reach expression levels comparable to OAS1-p46. Thus, the effects of genetic variants on OAS1 expression can be compensated by IFN treatment to overcome impaired viral clearance leading to severe COVID-19.