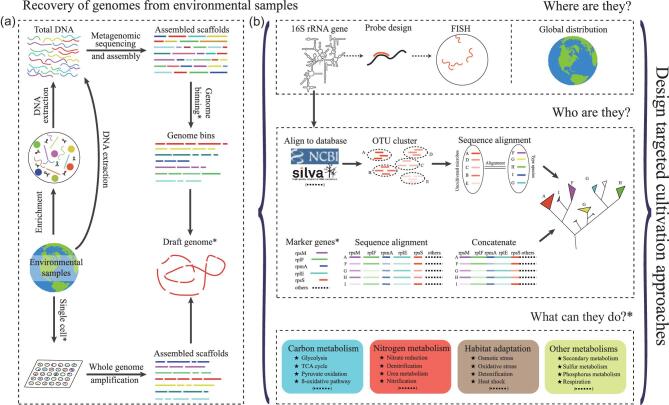
Figure 1.
Schematic of research approaches to answer the basic scientific questions about MDM. (a) Typical pipeline to produce SAGs or MAGs. Single-cell isolation requires specialized instrumentation, such as flow cytometry, microfluidics or micromanipulators, and clean rooms are also required for downstream handling. Genome binning tools are available, such as MetaBAT, MaxBin, GroopM, MetaWatt and CONCOCT. The quality of draft genomes could follow the minimum standards information about SAGs and MAGs, which was proposed by the Genome Standards Consortium to facilitate communication and more robust comparative genomic analyses of microbial diversity [3]. (b) Downstream analysis based on MDM genomes and a flowchart illustrating how genomic data can be integrated. Marker genes used for phylogenetic analysis are flexible enough to accommodate changes over time. Combination of multi-omics, FISH, MAR, NanoSIMS, Raman and other techniques would provide compelling evidence for microbial function and the framework to design targeted cultivation strategies.