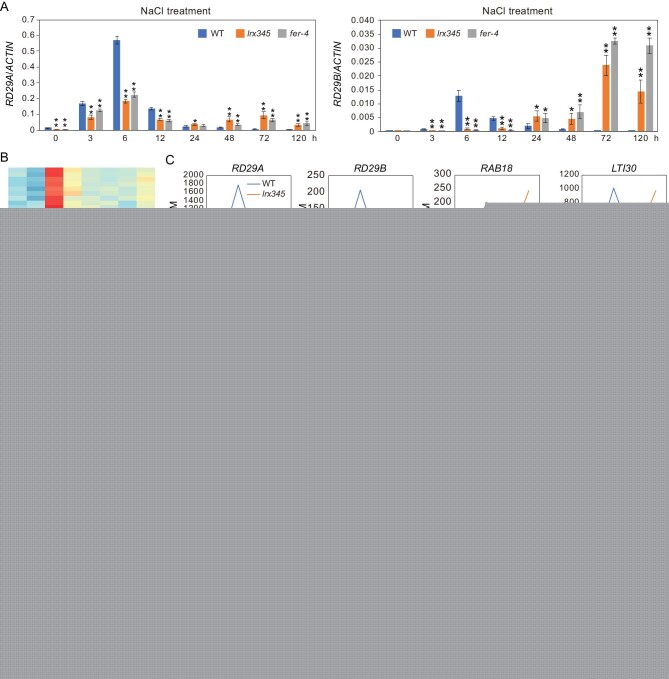
Figure 5.
LRX3/4/5 and FER are required for the dynamic regulation of salt stress-responsive gene expression. (A) Ten-day-old seedlings of the wild type, lrx345 and fer-4 were treated with NaCl for 0, 3, 6, 12, 24, 48, 72 and 120 h. The transcript levels of RD29A and RD29B were analyzed by qRT-PCR. ACTIN8 was used as an internal control. (B) RNA-seq assay was performed for the wild type and lrx345 after NaCl treatment for 0, 6, 24 and 72 h. Heat map shows the genes that exhibited lower transcript levels after NaCl treatment for 6 h, and showed higher transcript levels after treatment for 72 h in the lrx345 mutant compared with that in the wild type. (C) Representative genes that display similar expression patterns as RD29A and RD29B after salt treatment for 0, 6, 24 and 72 h. The transcript profile of each gene in the wild type and lrx345 mutant was generated from RNA-seq data. (D) Wild type seedlings were pretreated with or without EGCG (100 μM) for 2 h before being subjected to NaCl treatment for 0, 6, 24 and 72 h. Transcript levels of RD29A, RD29B and RAB18 were analyzed. ACTIN8 was used as an internal control. Values in (A) and (D) are means of three biological replicates ± SD, *P < 0.05 and **P < 0.01 (Student's t-test).