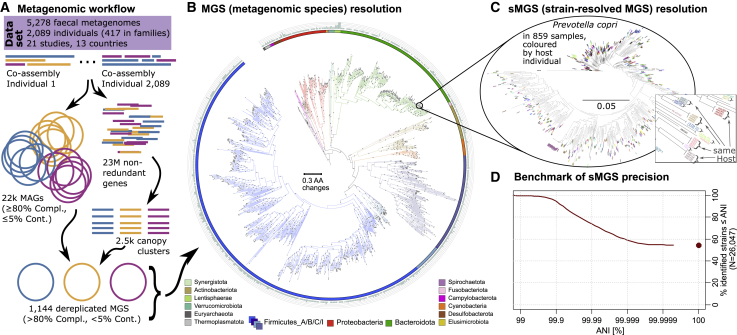
Figure 1.
Bioinformatic workflow leading to strain-resolved metagenomic species
(A) 5,278 longitudinal metagenomes were co-assembled per individual host (n = 2,089) (Figure S1). From these co-assemblies, a gene catalog with 23,137,742 genes was created and used to cluster 2,474 canopies. In parallel, metagenomic assembled genomes (MAGs) were calculated from the co-assemblies. MAGs and canopy clusters were combined and dereplicated to 1,144 high quality (>80% completeness, <5% contamination) MGS.
(B) Phylogeny and taxonomic assignment of all 1,144 MGS. The outer circle indicates missing taxonomic assignment levels (species, genus, family, order, and class), all MGS had at least phylum-level assignments, 83% were named at the genus level. Branches with >90 bootstrap support have gray circles.
(C) Intraspecific phylogeny exemplified for Prevotella copri. 859 sMGS were reconstructed from 859 metagenomic samples with ≥2X P. copri coverage, tree tips are randomly colored by the host individual. Monophyletic sMGS within the same host or host family were used to identify strains persisting in individuals or families.
(D) Identified strains were used to benchmark sMGS precision. The average nucleotide identity (ANI) was calculated between genetic sequences of strains found recurrently in individuals or families, using core genes of a species (see STAR Methods). 55% of these sequences were completely identical (100% ANI), with 95% of strains having <99.9% ANI in their representative sequences. For brevity, MGS and sMGS will be referred to as species and strains, respectively, in the main text. MAG, metagenomic assembled genome; compl., Cont, completeness and contamination of genomic bin; MGS, metagenomic species; sMGS, strain-delineated MGS; ANI, average nucleotide identity.