Figure 5:
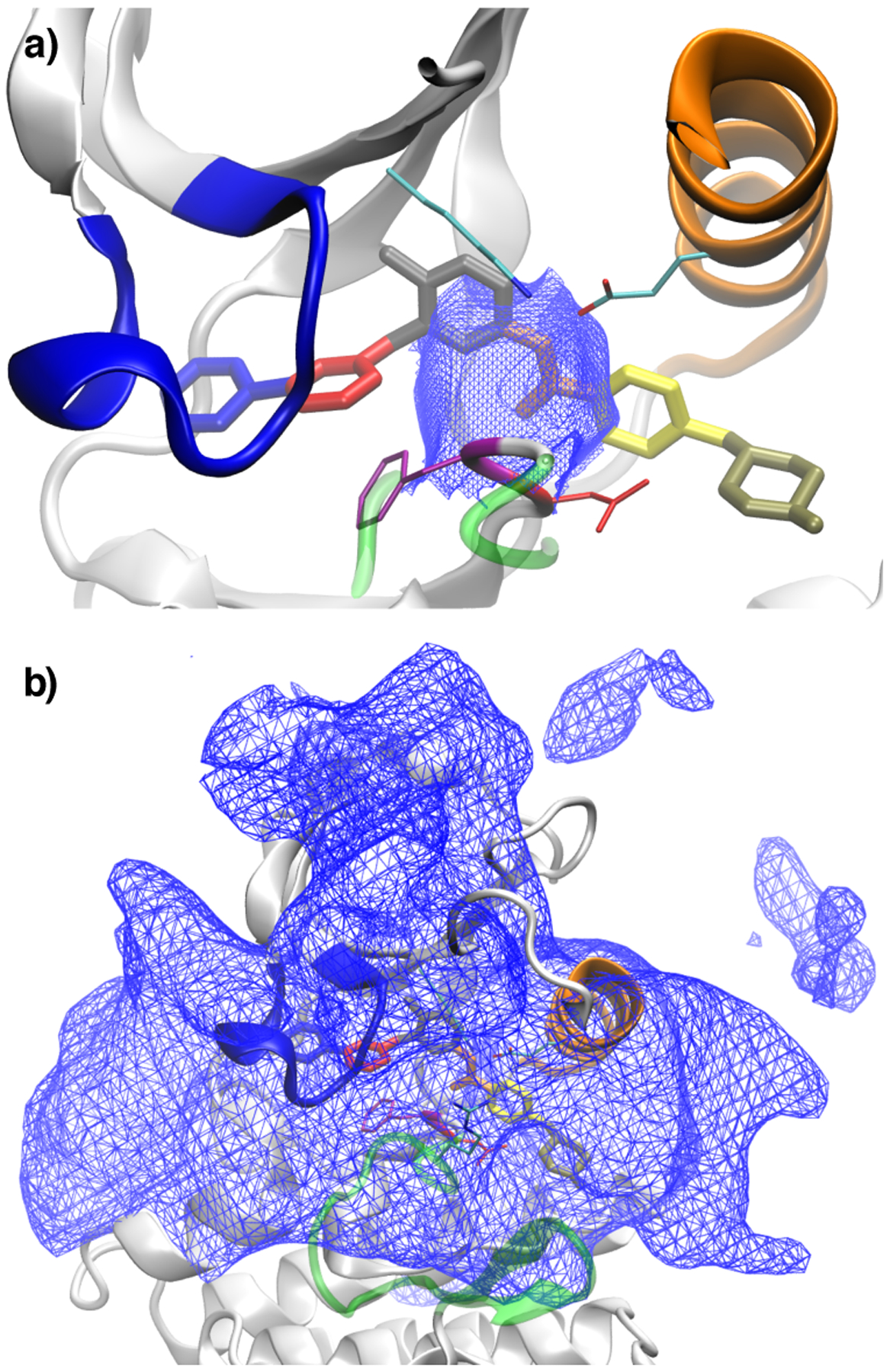
Dividing surfaces in (imatinib) center-of-mass feature space between different classes of trajectories: (a) shows the surface between trajectories that rebind to the crystallographic mode and trajectories that are trapped in intermediate states; (b) shows the dividing surface between trajectories trapped in intermediate states and trajectories that lead to full dissociation. The protein and ligand conformation shown in both figures corresponds to the crystal structure 2HYY. The two surfaces in (a) and (b) delineate the rugged and complex intermediate region of Abl kinase where are located several non-specific long-lived associated states of imatinib. See Theory and Method for additional details on how the surfaces were generated.
