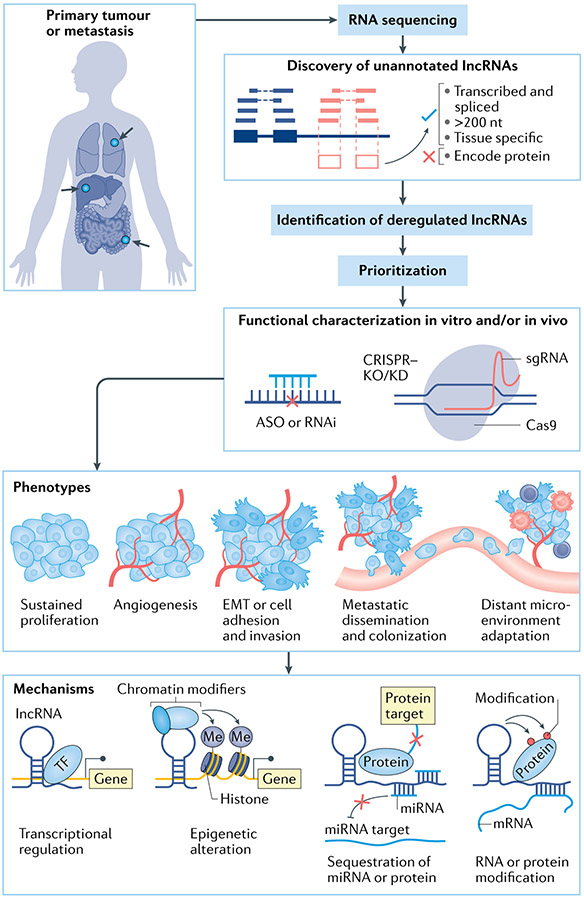
Fig. 1 ∣. Long noncoding RNAs in cancer.
Starting from discovery to characterization of their functions and mechanisms. First, tumour specimens and normal tissues sampled from patients with cancer are subject to RNA sequencing. Sequencing reads are then aligned to the reference genome, which allows quantification of RNA abundance and discovery of unannotated long noncoding RNAs (lncRNAs). Comparisons of transcript abundance between tumour and normal and/or between metastasis and primary tumour allow identification of deregulated lncRNAs. Subsequent omics data integration, computational analysis and high-throughput functional screening using techniques such as CRISPRi allow prioritization of lncRNA candidates for further in-depth functional and mechanistic characterization. Perturbation and manipulation of the lncRNAs can be performed via antisense oligonucleotide (ASO), small interfering RNA (siRNA), CRISPR-based methods in vitro and in vivo to generate phenotypic changes that can be observed and evaluated. LncRNAs are known to have both oncogene and tumour suppressor roles and contribute to tumour progression and metastasis. Mechanistically, lncRNAs can regulate the expression of other genes by interacting with other molecules resulting in transcriptional regulation, epigenetic alteration, sequestration of RNA or protein and enhancer regulation. EMT, epithelial–mesenchymal transition; KO/KD, knockout/knock-down; Me, methyl; miRNA, microRNA; RNAi, RNA interference; sgRNA, single guide RNA; TF, transcription factor.