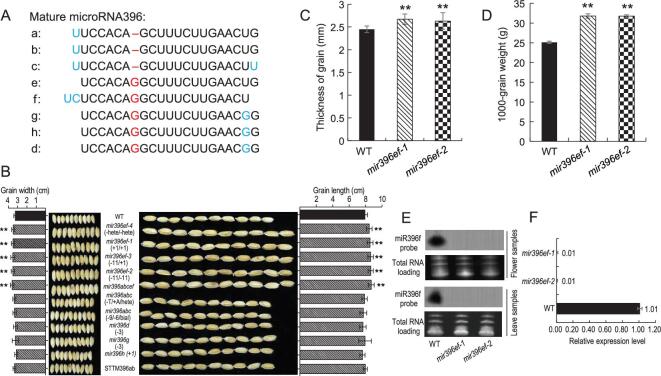
Figure 1.
miR396ef isoforms regulate rice-grain development (A) miR396 members in rice. Isoform differences are highlighted in blue. Red color indicates the difference at position seven. (B) Comparison of the 10-grain width and 10-grain length between mir396 mutant plants and wild-type (WT). The genotypes are labeled after the name of mutants in the brackets. Plus (+) and minus (−) and subsequent number represent the nucleotides inserted and deleted at the sgRNA target sites, and mutations on different mir396 members are separated by slashes. The ‘hete’ means heterozygous mutation on the corresponding members. The ‘bial’ means biallelic mutation on the corresponding member. (C) The grain thickness of mir396ef and WT plants. (D) The 1000-grain weight of mir396ef and WT plants. (E) Northern blotting detection of miR396ef in flowers and leaf samples of WT and mir396ef plants. Total RNA stained with ethidium bromide was used as a loading control. (F) Stem-loop RT-qPCR of miR396ef in WT and mir396ef leaf samples. U6 was used to normalize samples. Data are presented as means ± SD (n = 100 in (B)–(D); n = 3 in (F)). P-values (versus the WT) were calculated with Student’s t-test, two-tailed. **P < 0.01.