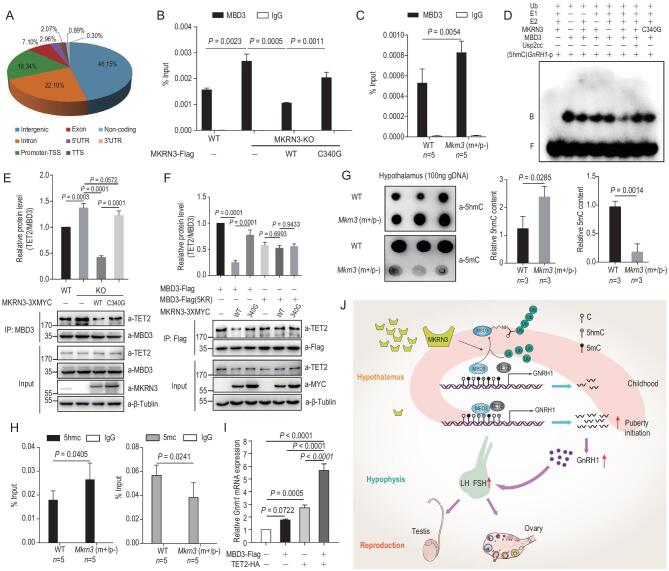
Figure 6.
MKRN3–MBD3–TET2 axis regulates the methylation status of mammalian genome, including GNRH1 promoter. (A) ChIP-seq analysis identified MBD3-binding regions in the human genome. Wild-type HEK293T cells were subjected to ChIP-seq analysis, using anti-MBD3 to enrich DNA-fragment complexes with endogenous MBD3. Percentages shown in the chart indicate the relative abundance of MBD3-bound regions in different structural parts of the genome. (B) MKRN3 inhibits the binding of endogenous MBD3 to the GNRH1 promoter in a manner depending on its E3 ligase activity. ChIP-qPCR assays were performed with HEK293T cells, wild-type or MKRN3−/− that were transfected with or without wild-type or C340G mutant MKRN3. Data are presented as mean ± SD, one-way ANOVA, with Bonferroni post-hoc test, three independent samples in each group. (C) Loss of Mkrn3 promotes the binding of endogenous MBD3 to the Gnrh1 promoter in primarily dissected neurons from mouse hypothalamus at postnatal day 15. ChIP-qPCR assays were performed in primarily dissected neurons from the hypothalamus of wild-type and Mkrn3 (m+/p−) mice. Data are presented as mean ± SD, two-tailed unpaired t-test, five samples in each group. (D) MKRN3-mediated ubiquitination compromised the affinity of recombinant MBD3 to 5hmC (5-hydroxymethylcytosine)-containing DNA sequences derived from the human GNRH1 promoter. EMSA was performed using −1716 to −1541 bp of the GNRH1 promoter with all cytosines substituted by 5hmC as a probe, labeled with γ-32P. Recombinant MBD3 was subjected to in vitro ubiquitination under varying conditions, then immunoprecipitated with anti-Flag beads for purification before using in the EMSA assay. B, bound probe; F, free probe. (E) MKRN3 impairs MBD3–TET2 interaction in HEK293T cells. Wild-type or MKRN3−/− cells were transfected with Myc-tagged wild-type or C340G mutant MKRN3; 48 hrs later, cell lysates were immunoprecipitated with anti-MBD3 antibody, and TET2 and other proteins as indicated were detected by immunoblotting. Data are presented as mean ± SD, one-way ANOVA, with Bonferroni post-hoc test, three independent experiments. (F) MKRN3-mediated ubiquitination disrupts MBD3–TET2 interaction in HEK293T cells. MKRN3/MBD3 double knockout (MKRN3−/−MBD3−/−) HEK293T cells were co-transfected with either wild-type or C340 mutant MKRN3-3xMyc and Flag-tagged wild-type or 5KR mutant MBD3; 48 hrs later, cell lysates were immunoprecipitated with anti-Flag beads and subjected to immunoblotting with indicated antibodies. Data are presented as mean ± SD, one-way ANOVA, with Bonferroni post-hoc test, three independent experiments. (G) Genetic ablation of Mkrn3 reduced 5mC content and increased 5hmC content in mouse hypothalamic genomic DNA. A dot-blotting assay was performed with genomic DNA extracted from the hypothalamus of wild-type or Mkrn3 (m+/p−) mice at postnatal day 15, using anti-5mC or anti-5hmC antibodies. For each sample, 100 ng genomic DNA was loaded onto the nitrocellulose membrane. Three mice in each group, two-tailed unpaired t-test. (H) Mkrn3 deficiency was associated with increased 5hmC and decreased 5mC in the Gnrh1 promoter of the mouse hypothalamus. Genomic DNA was isolated from hypothalamus of wild-type and Mkrn3 (m+/p−) mice at postnatal day 15 and immunoprecipitated with anti-5hmC or 5mC antibodies, followed by qPCR using primers for the Gnrh1 promoter. Three mice in each group, two-tailed unpaired t-test. (I) MBD3 and TET2 synergistically activate Gnrh1 transcription. GT1-7 cells were transfected with MBD3, TET2 or both followed by qPCR analysis of Gnrh1 expression. GAPDH was taken as the endogenous control, data are presented as mean ± SD, one-way ANOVA, with Bonferroni post-hoc test, three independent experiments. (J) A model depicting how the MKRN3–MBD3–TET2 axis might regulate the timing of mammalian puberty.