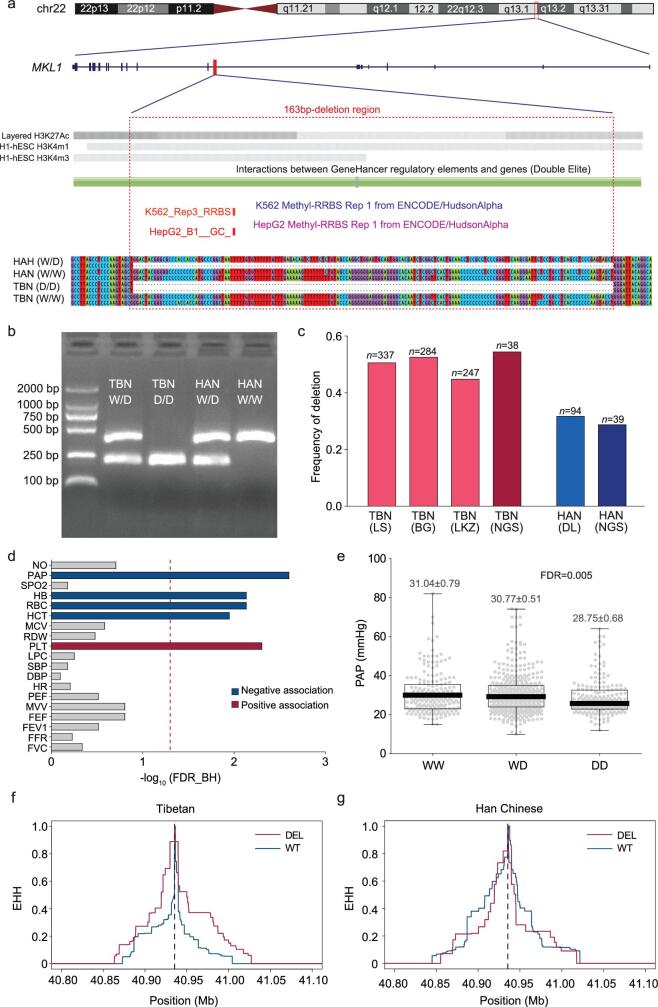
Figure 3.
The Tibetan-enriched MKL1 163-bp deletion and its association with physiological traits. (a) The schematic map indicating the genomic location (upper panel), epigenetic signals (histone modification and DNA methylation, middle panel) and sequence alignment (bottom panel) of the MKL1 deletion and its flanking sequences in Tibetans and Han Chinese. (b) Genotyping electromorphic of the MKL1 deletion. The two alleles are indicated as ‘W’ (wild type) and ‘D’ (deletion). (c) Allele frequencies of the MKL1 deletion in Tibetans (TBN) and Han Chinese (HAN); TBN (LS), Tibetans at Lhasa; TBN (BG), Tibetans at Bange; TBN (LKZ), Tibetans at Langkazi; HAN (DL), Han Chinese at Dalian; TBN (NGS) and HAN (NGS), Tibetans and Han Chinese from the NGS data (Methods). (d) Genetic association between the MKL1 deletion and multiple physiological traits in Tibetans (n = 868). The dot line in red refers to the cut-off of statistical significance with false discovery rate (FDR) of ∼5% by Benjamini and Hochberg [42] and the trait abbreviations are described in Methods. (e) Comparison of pulmonary arterial pressure levels among three different genotypes at the MKL1 deletion; W-wide type (non-deletion). P-value was calculated assuming an additive model with multiple-testing correction using Bejamini and Hochberg FDR control (FDR_BH) (Methods). (f) and (g) Estimation of EHH decay of haplotypes in Tibetans (f) and Han Chinese (g) surrounding the MKL1 163-bp deletion. The physical position of the 163-bp deletion is indicated by the vertical dashed line in (f) and (g).