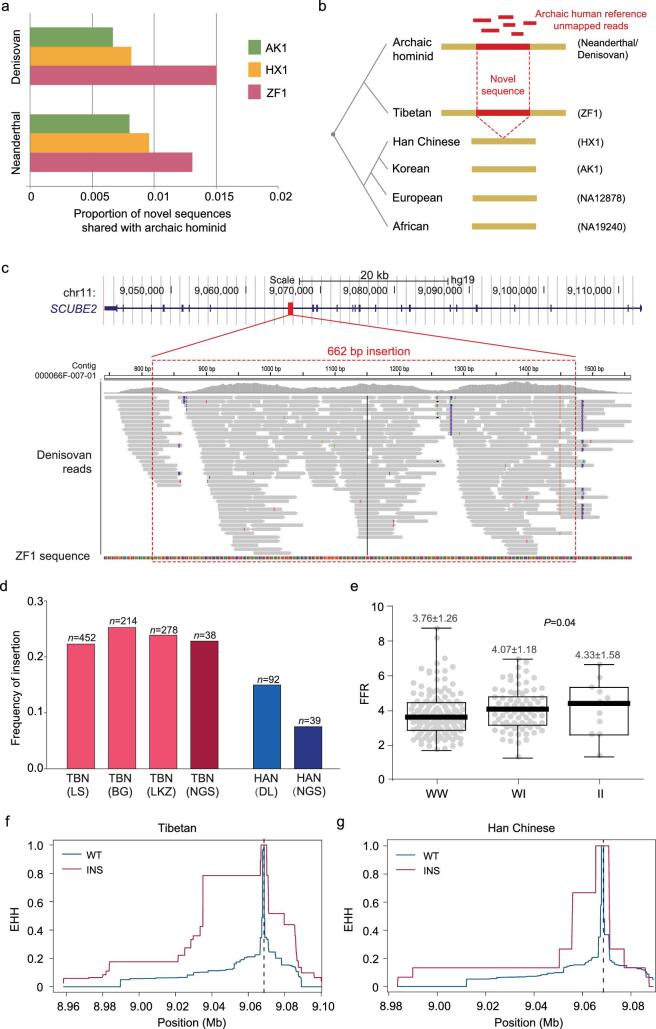
Figure 4.
ZF1-speicific novel sequences shared with archaic hominins. (a) Genomic proportion of the non-reference sequences shared with archaic hominids in the three de novo assembled Asian genomes. (b) Schematic diagram of the ZF1-sepcific non-reference sequences shared with archaic hominids. Novel non-reference sequences (red) shared between Tibetan (ZF1) and archaic hominids but absent in the other representative modern human genomes (one European, one African and two Asian genomes). (c) A novel sequence shared between ZF1 and archaic hominid in the intron of SCUBE2. This sequence was found in both Denisovan and Altai Neanderthal. Shown here is the Denisovan reads mapping. The upper panel shows the position of this 662-bp novel sequence in the human reference genome GRCh37. The bottom panels indicate the Denisovan reference unmapped reads aligned to the ZF1 contig. (d) Allele frequencies of the 662-bp insertion in Tibetans (TBN) and Han Chinese (HAN). TBN (LS), Tibetans at Lhasa; TBN (BG), Tibetans at Bange; TBN (LKZ), Tibetans at Langkazi; HAN (DL), Han Chinese at Dalian; TBN (NGS) and HAN (NGS), insertion frequency estimated by next-generation-sequencing data (Methods). (e) Comparison of the FEV1/FVC ratios among three different genotypes at the SCUBE2 insertion; I, insertion; W, wide type (non-insertion). (f) and (g) Estimation of EHH in TBN (f) and HAN (g) around the SCUBE2 insertion at Chr11:9068607. The physical position of the insertion is indicated by the vertical dashed line in (f) and (g).