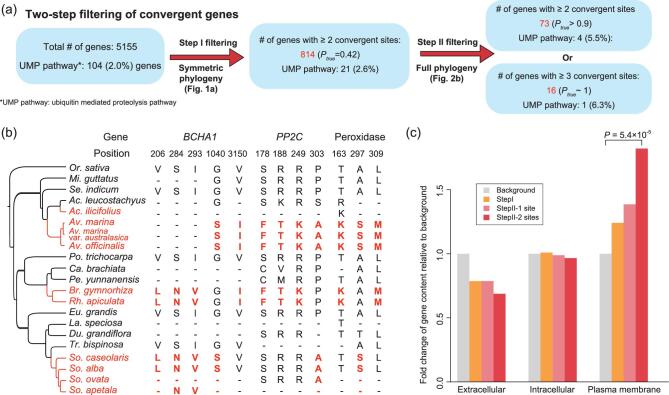
Figure 2.
Genic convergence among mangroves. (a) A two-step procedure for identifying genes of convergence. In Step I, 814 candidate genes are identified based on the small phylogeny of Fig. 1a. A modest probability of true convergence (Ptrue = 0.42) is attained. In step II using the full phylogeny of panel (b), further screening of the 814 genes yields 73 genes of high confidence. Genes of the ‘ubiquitin-mediated proteolysis’ pathway are enriched in the sets. (b) Three examples of convergent genes in mangroves are shown. These genes have at least three convergence sites and are associated with salinity tolerance. Red coloring in the phylogenetic tree is used for mangrove species. (c) Proteins of convergence are enriched on plasma membrane. Subcellular localization is classified as extracellular, intracellular or membrane-bound based on the prediction of CELLO [47]. In each class, the percentage of the 5155 background genes is normalized as 1 while the percentages for the convergent genes are shown as the fold change relative to the background. For genes producing membrane-bound proteins, the enrichment of convergent genes, relative to the background, increases as the criteria become more stringent (P = 5.4 × 10−5 by Fisher's exact test). Such a pattern is not observed for extracellular and intracellular proteins.