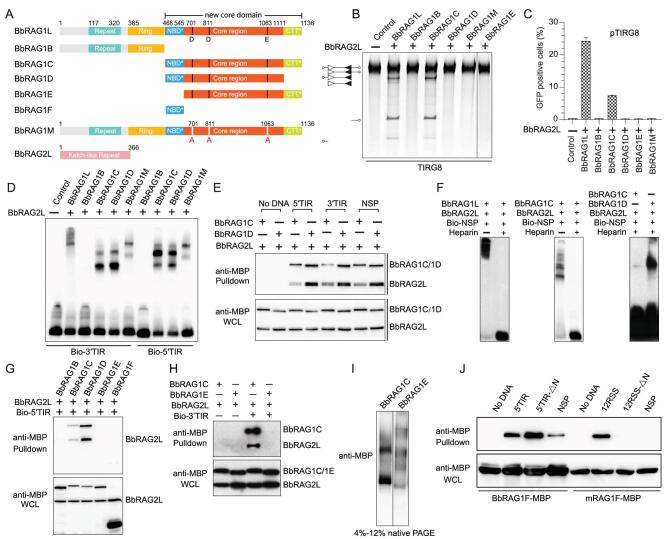
Figure 6.
Domains in BbRAG1L essential for the activity of ProtoRAG. (A) Diagram showing the domains of truncated BbRAG1Ls and the active site mutations in BbRAG1L and BbRAG2L. Domains are defined according to the sequence alignment with mouse RAG1 recombinase [20]. (B) Cleavage of the TIRG8 substrate by the indicated BbRAGL proteins in vitro. The composition of the cleavage product is shown on the left according to the lengths of the corresponding fragments. Unfilled and filled triangles indicate the 5′TIR and 3′TIR sequences of ProtoRAG, respectively. (C) Quantification of the GFP-positive cells produced by BbRAGL-mediated recombination with the pTIRG8 substrate. The composition of the distinct truncated BbRAG1L proteins is shown in (A). (D) EMSA assay to detect the binding of BbRAGL proteins with 5′TIR and 3′TIR. The indicated BbRAGL proteins and their truncated forms were purified. The binding reactions were separated on a 3.5/8% native TBE-PAGE gel. (E) Pull-down assay to detect the binding of BbRAG1C and BbRAG1D with 5′TIR and 3′TIR. Both BbRAG1C and BbRAG1D were co-expressed with BbRAG2L. The NSP probe was mutated from 5′TIR by scrambling the sequences. (F) EMSA assay to detect the binding of BbRAG1L, BbRAG1C and BbRAG1D with nonspecific DNA. The NSP probe was mutated from 5′TIR by scrambling the sequences. (G) Pull-down assay to detect the binding of BbRAG1E and BbRAG1F with 5′TIR. Both BbRAG1E and BbRAG1F were co-expressed with BbRAG2L. WCL, whole-cell lysates. (H) Pull-down assay to detect the binding of BbRAG1E/BbRAG2L with 3′TIR. (I) Electrophoretic separation of singly purified BbRAG1C and BbRAG1E by 4%–12% native PAGE. (J) Comparison of the DNA-binding ability of the NBD* domain of BbRAG1 (BbRAG1F) and the NBD domain of mRAG1 (mRAG1F). The DNA probes are shown as indicated and the TR5 in 5′TIR-ΔN and the nonamer in 12RSS-ΔN were mutated. The NSP probe was mutated from 5′TIR and 12RSS by scrambling the TIR and 12RSS sequences, respectively.