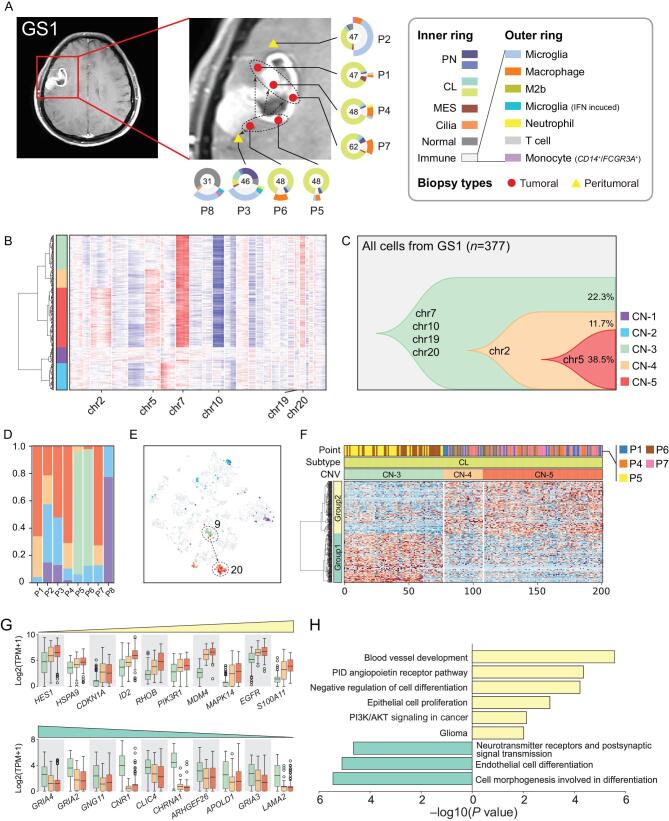
Figure 4.
Glioma clonal evolution of Patient GS1 at spatial and temporal resolution. (A) MRI image of Patient GS1. Yellow and red markers in zoomed image represent peritumoral and tumoral sampling points. Ring plot in the right and bottom displays cell components of each point. Color of the inner ring shows classified glial cell subtypes, and the outer ring shows detailed immune cell subtypes. Cell numbers are labeled in the center of these ring plots. (B) Single-cell CNV heatmap. Cells were divided into five groups by hierarchical clustering. (C) Clonal evolution trail followed by accumulating CNV events. Each color represents a CNV subclone and chromosomes are labeled in which copy number alteration occurred during clonal transition. (D) CNV subclone components in each sampling point, P5/6 had a different component compared with other tumoral/peritumoral points. (E) CNV subclone distributions in t-SNE coordinates. When CN-3 developed into CN-4/5, cells translocated from cluster 9 to cluster 20. (F) Heatmap of differentially expressed genes between CN-3/4/5 CL glial cells. Differential genes could be divided into two gene-sets, marked with the left color bar. (G) Box plot of differential genes. The increased (upper) and decreased (lower) gene expression profiles followed CNV accumulation. Boxes were colored by CNV subgroup. (H) Functional enrichment of increased and decreased gene-sets.