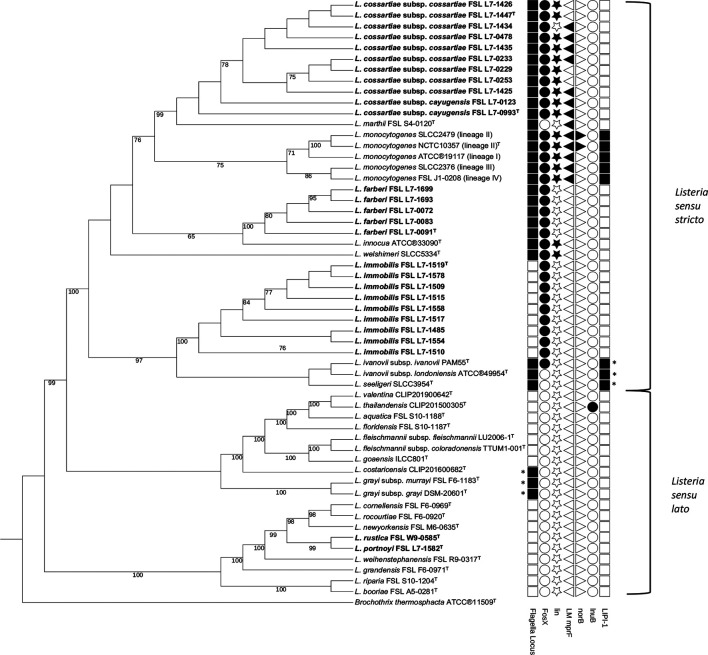
Fig. 2.
Maximum-likelihood consensus phylogeny based on the concatenation of a 120-bacterial protein marker set (bac120) from GTDB-Tk analysis of the 27 draft genomes representing the five novel Listeria species and two subspecies and the same reference set used for ANIb analysis (see Fig. 1). The phylogeny was reconstructed using RAxML v8.2.12 and the PROTGAMMAILGF model. The values on the branches represent bootstrap values based on 1000 replicates; bootstrap values <70 are not shown. The tree is rooted at the midpoint and includes the outgroup Brochothrix thermosphacta ATCC®11509T. The tree was edited with iTOL v5 [43]. Novel Listeria species are bolded and type strains are identified with a superscript T. The presence/absence of key genes, operons, and loci in the draft genomes were mapped onto the tree using iTOL [43] and indicated with filled/unfilled symbols (filled indicates presence, unfilled indicates absence). Symbols with an ‘*’indicate loci containing diversified genes and consequently only some genes were detected using a blastn search with reference L. monocytogenes genes from the PasteurMLST databases [61, 62]; specifically (i) not all genes in the flagella locus sourced from the cgMLST1748 database (lmo0676 thru lmo0717 [76]; Table S4) were detected in the L. grayi and L. costaricensis genomes, and (ii) not all LIPI-1 genes sourced from the Virulence database were detected in the L. ivanovii and L. seeligeri genomes. Detection of diversified flagella and LIPI-1 genes was achieved via alternative search methods including (i) locating the genes in the NCBI GenBank annotated genomes, or (ii) using more closely related reference genes (e.g. the L. ivanovii prfA gene cluster).