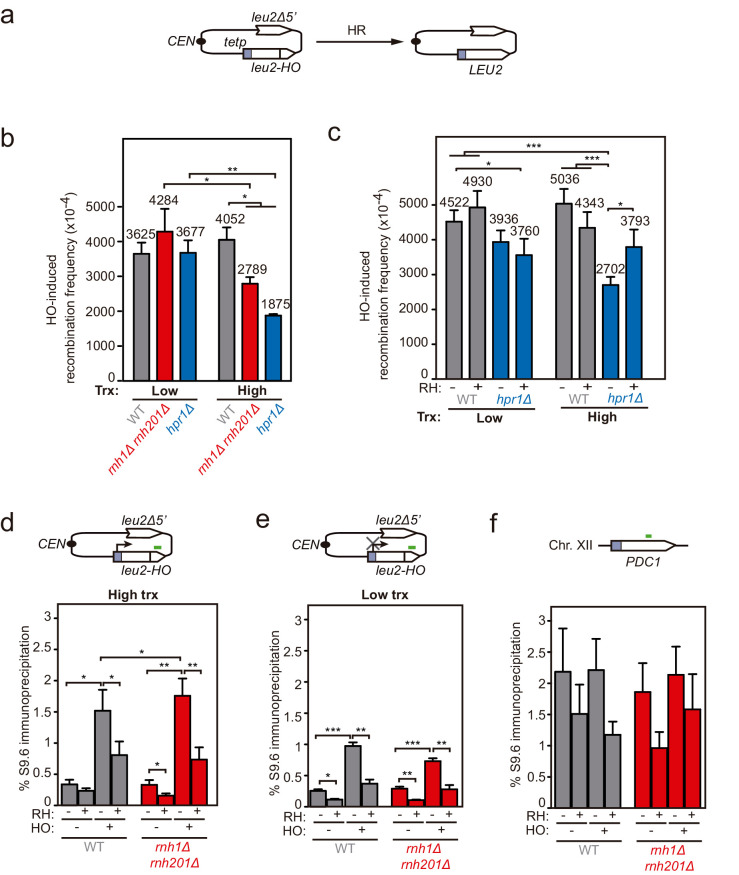
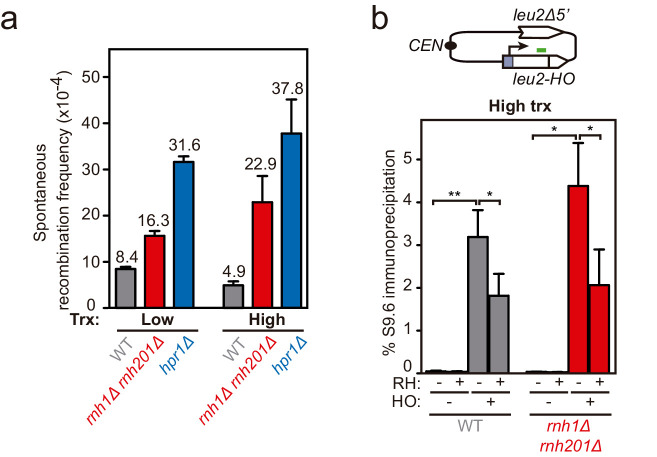
Figure 5. Genetic analysis of the repair and DNA-RNA hybrid accumulation at endonuclease-induced DSBs.
(a) Scheme of the pCM189-L2FHO recombination system. (b) Frequency of HO-induced recombination in wild type (WS), rnh1Δ rnh201Δ (WSR1R2) and hpr1Δ (WSHPR1) strains transformed with pTINV-FHO under low or high transcription (n≥3). (c) Frequency of HO-induced recombination in wild type (WS) and hpr1Δ (WSHPR1) strains transformed with either pRS314 (RH−) or pRS314-GALRNH1 (RH+) and pTINV-FHO under low or high transcription (n=9). (d) DRIP with the S9.6 antibody in the leu2-HO allele as depicted on top and in either spontaneous conditions (HO−) or after HO induction (HO+) in wild type (WS) and rnh1Δ rnh201Δ (WSR1R2) strains transformed with pTINV-FHO under high transcription and either non-treated (RH−) or after in vitro RNase H treatment (RH+) (n=5). (e) DRIP with the S9.6 antibody in the leu2-HO allele as depicted on top and in either spontaneous conditions (HO−) or after HO induction (HO+) in wild type (WS) and rnh1Δ rnh201Δ (WSR1R2) strains transformed with pTINV-FHO under low transcription and either non-treated (RH−) or after in vitro RNase H treatment (RH+) (n=4). (f) DRIP with the S9.6 antibody in the PDC1 gene as depicted on top and in either spontaneous conditions (HO−) or after HO induction (HO+) in wild type (WS) and rnh1Δ rnh201Δ (WSR1R2) strains transformed with pTINV-FHO under high transcription and either untreated (RH−) or after in vitro RNase H treatment (RH+) (n=5). Mean and SEM of independent experiments consisting in the median value of six independent colonies each are plotted in (b–f) panels. *p≤0.05; **p≤0.01; ***p≤0.001 (unpaired Student’s t-test in (b) panel and paired Student’s t-test in (c–f) panels). See also Figure 5—figure supplement 1. Data underlying this figure are provided as Figure 5—source data 1. DRIP, DNA-RNA immunoprecipitation; HR, homologous recombination; trx, transcription.